For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LALLDRTAALRHGADVVHRRLGADLAAGANRIAEDDTMEVVAVVGGRTEELNGLVIGPQEGLGGQAAVLR RTVAVHDYCMSRDITHDFDHAVRAEGLRAAMVAPIVRAHRLYGLLYAARRSPVAWTDRDRAELLSLARQA AVAMEVADSAREMAEVAAFGERQRLAIRLHDSVGATLFSLRVALMSAQEETEPGRREELLSDALVLTERA TSELRAQTLSLHDLPEDKALAVTLEGDCRDFATRTGTAAELVVLGDLPLLDSARTDALRRTAREALLNVE KHAGAHSVVVSLYRAEDGVGLTVADDGTAPDIASEHRRGLGLRATVERIERVGGWLSCTANEEGGGTVQA WVPAARAFRPVGP*
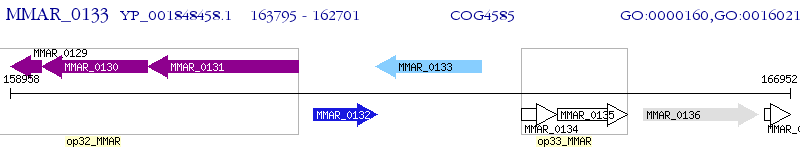
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0133 | - | - | 100% (364) | two-component sensor histidine kinase |
| M. marinum M | MMAR_1517 | devS | 3e-17 | 25.59% (254) | two-component sensor histidine kinase DevS |
| M. marinum M | MMAR_3479 | - | 3e-13 | 26.06% (330) | two-component sensor histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3156c | devS | 1e-18 | 27.17% (254) | two component sensor histidine kinase DEVS |
| M. gilvum PYR-GCK | Mflv_3844 | - | 1e-13 | 26.56% (256) | GAF sensor signal transduction histidine kinase |
| M. tuberculosis H37Rv | Rv3132c | devS | 1e-18 | 27.17% (254) | two component sensor histidine kinase DEVS |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3890c | - | 1e-18 | 26.80% (362) | histidine kinase response regulator |
| M. avium 104 | MAV_5281 | - | 4e-13 | 28.29% (258) | histidine kinase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_5241 | - | 3e-18 | 27.95% (254) | GAF family protein |
| M. thermoresistible (build 8) | TH_0388 | - | 1e-13 | 25.22% (230) | TWO COMPONENT SENSOR HISTIDINE KINASE DEVS |
| M. ulcerans Agy99 | MUL_2422 | devS | 3e-17 | 25.59% (254) | two component sensor histidine kinase DevS |
| M. vanbaalenii PYR-1 | Mvan_1413 | - | 2e-14 | 26.62% (278) | GAF sensor signal transduction histidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3156c|M.bovis_AF2122/97 -------MTTGGLVDENDGAAMRPLRHTLSQLRLHELLVEVQDRVEQIVE
Rv3132c|M.tuberculosis_H37Rv -------MTTGGLVDENDGAAMRPLRHTLSQLRLHELLVEVQDRVEQIVE
MUL_2422|M.ulcerans_Agy99 MTDEPHPQDAGPSLGVPSGDGLRPLRLTLSQLRLRELLVEVQERVEQIVE
Mflv_3844|M.gilvum_PYR-GCK ----MHDGAVADADDHRARDGARPVRTTLSQLRLRELLTEVQDRVEQIIR
MSMEG_5241|M.smegmatis_MC2_155 ------MGGVNGHHPRRDDDQRTPLRDTLSQLRLRELLSEVQDRVEQIVE
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 -------------MVEDVGAQPNRVTAELAHLQLRELLAEVQGRIEQIVD
Mvan_1413|M.vanbaalenii_PYR-1 ---------------MSNRQQRAFGRRHPADLRR------IHEQLDELAA
MMAR_0133|M.marinum_M --------------------------------------------------
MAV_5281|M.avium_104 --------------------------------------------------
Mb3156c|M.bovis_AF2122/97 G-RDRLDGLVEAMLVVTAGLDLEATLRAIVHSATSLVDARYGAMEVHDRQ
Rv3132c|M.tuberculosis_H37Rv G-RDRLDGLVEAMLVVTAGLDLEATLRAIVHSATSLVDARYGAMEVHDRQ
MUL_2422|M.ulcerans_Agy99 G-RDRLDGLVEAMLVVTSGLELDATLRTIVHSATNLVDARYCALEVHDRD
Mflv_3844|M.gilvum_PYR-GCK G-RDRLDGLVEAMLAVTSGLELDETLRTIVHTAIELVDAQYGALGVRGHD
MSMEG_5241|M.smegmatis_MC2_155 G-RDRLDGLVEAMLVVTSGLELDVTLRTIVHTAIELVDARYGALGVRGED
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 GTRGRMDALLDAVMAVSSGLELDATLREIVCAASELVSARYGALGVVGSD
Mvan_1413|M.vanbaalenii_PYR-1 D-RDQMGQLLQVAVEIGSDLELNATLQRIVRAARGMTAARYGAIGVWGTD
MMAR_0133|M.marinum_M --------------------------------------------------
MAV_5281|M.avium_104 --------------------------------------------------
Mb3156c|M.bovis_AF2122/97 HRVLHFVYEGIDEETVRRIGHLPKGLGVIGLLIEDPKPLRLDDVSAHPAS
Rv3132c|M.tuberculosis_H37Rv HRVLHFVYEGIDEETVRRIGHLPKGLGVIGLLIEDPKPLRLDDVSAHPAS
MUL_2422|M.ulcerans_Agy99 KRVLQFVYEGIDEDTVARIGHLPEGLGIIGLLIDEPKPLRLEDISQHPAS
Mflv_3844|M.gilvum_PYR-GCK QELVEFIYEGIDEPMRERIGHLPEGRGVLGVLINDPKPIRLEDISRHADS
MSMEG_5241|M.smegmatis_MC2_155 HHLIEFIYEGITPEVREEIGPLPEGRGVLGVLIDDPKPIRLDDIRLHPAS
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 EKLAEFVYEGIDEATRDLIGPLPTGHGVLGVVIDEAKPLRLRDIAAHPAS
Mvan_1413|M.vanbaalenii_PYR-1 DLLSDFVHDGMDEGTVQAVGHPPVGKGLLGFLRQGTDPVRLDNAAEHPSA
MMAR_0133|M.marinum_M ------MLALLDRTAALRHGADVVHRRLGADLAAGANRIAEDDTMEVVAV
MAV_5281|M.avium_104 --MLRAAICHLREQLRRRGELIPVGLTWASLIAVDAALLLGGTIGTLQR-
Mb3156c|M.bovis_AF2122/97 IGFPPYHPPMRTFLGVPVRVRDESFGTLYLTDKTNGQPFSDDDEVLVQAL
Rv3132c|M.tuberculosis_H37Rv IGFPPYHPPMRTFLGVPVRVRDESFGTLYLTDKTNGQPFSDDDEVLVQAL
MUL_2422|M.ulcerans_Agy99 IGFPSHHPPMRTFLGVPVRVRDVSLGTLYLTDKTNGQPFSDDDELLVQAL
Mflv_3844|M.gilvum_PYR-GCK VGFPPNHPPMRTFLGVPVRIRDEVFGNLYLTEKSDGQPFSEDDEVLVQAL
MSMEG_5241|M.smegmatis_MC2_155 VGFPPNHPPMRTFLGVPVRTRDKVFGNLYLTEKTNGQPFSEDDEVLVQAL
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 VGFPANHPPMRTFLGVPVQVRGEVFGRLYLTEKLDGQEFTEDDEVVVRAL
Mvan_1413|M.vanbaalenii_PYR-1 VGFPQHHPAMRAFIGVPIVIRGEPFGSLYLADDRPDFAFAEADEMTVRAL
MMAR_0133|M.marinum_M VGG-----------------RTEELNGLVIGPQ--------------EGL
MAV_5281|M.avium_104 ----------------PAADLPVSLTAFAIALSPTVAFFVFNMKMTPGPL
Mb3156c|M.bovis_AF2122/97 AAAAGIAVANARLYQQAKARQSWIEATRDIATELLSGTEP-ATVFRLVAA
Rv3132c|M.tuberculosis_H37Rv AAAAGIAVANARLYQQAKARQSWIEATRDIATELLSGTEP-ATVFRLVAA
MUL_2422|M.ulcerans_Agy99 AAAAGIAIANARLYEQAKARQAWIGATRDIATELLSGTEP-ASVFRLVAE
Mflv_3844|M.gilvum_PYR-GCK AAAAGIAIDNARLYAQSQARQSWIEATRDIGTELLSGIDP-AGVFRRVAD
MSMEG_5241|M.smegmatis_MC2_155 AAAAGIAVDNARLYEESQARQAWIAATRDIGTQLLSGTDP-ATVFRLVAA
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 AGAAGIAIENARLYQHARRREQWQRATADITAELLGGVDP-GKALHLVAS
Mvan_1413|M.vanbaalenii_PYR-1 ASIAAVAIDNARLLDHARATVRWTDAGREITAAVLGGDDPYLRPLQLIVG
MMAR_0133|M.marinum_M GGQAAVLRRTVAVHDYCMSRDITHDFDHAVRAEGLR--------------
MAV_5281|M.avium_104 WATWTASTALTLFVTSTPIRADFAPALLVLMVLVVATLAS----------
Mb3156c|M.bovis_AF2122/97 EALKLTAADAALVAVPVDEDMPAADVGELLVIETVGSAVASTVGRTIPVA
Rv3132c|M.tuberculosis_H37Rv EALKLTAADAALVAVPVDEDMPAADVGELLVIETVGSAVASIVGRTIPVA
MUL_2422|M.ulcerans_Agy99 EALKLTSADAALVAIPVDEDVPASEVAQLLVIETVGNAVAAAEGCTIAVA
Mflv_3844|M.gilvum_PYR-GCK RAQKLSGAAATLVAVPDDPDLPVGEVAELTVAASAGETIAAAHD-PIPVA
MSMEG_5241|M.smegmatis_MC2_155 EALTLTGADGTLVAVPADPDASAAEELVIVEVAGAVPAEVEAS--AIPVQ
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 SAALLAEADFAFIALPAGDEE---DTAELVVAVCEGKGAEVLNDKLIPID
Mvan_1413|M.vanbaalenii_PYR-1 RACELTGAEQGIVLVASDPEQPVDEVDTLVVSTAAGRYADDVLGQRVPVH
MMAR_0133|M.marinum_M --------------------------------------------------
MAV_5281|M.avium_104 --------------------------------------------------
Mb3156c|M.bovis_AF2122/97 GAVLREVFVNGIPRRVDRVDLEGLD-ELADAGPALLLPLRARGTVAGVVV
Rv3132c|M.tuberculosis_H37Rv GAVLREVFVNGIPRRVDRVDLEGLD-ELADAGPALLLPLRARGTVAGVVV
MUL_2422|M.ulcerans_Agy99 GTALAEVLLDSAPRQVDKIAVEDVD-ELGDMGPALVLPLRATDAVAGVVV
Mflv_3844|M.gilvum_PYR-GCK GTAIGDAFVHRTPGMTDRADIGIGA-TVG---PALLLPLRAPDAVPGVLI
MSMEG_5241|M.smegmatis_MC2_155 DNAIGQAFRDRAP---RRLDVLDGP-GLGG--PALVLPLRATDTVAGVLV
TH_0388|M.thermoresistible__bu -----------------------------------------------VLV
MAB_3890c|M.abscessus_ATCC_199 GSTSGSTFIDHEPRNVGRLAVNLAEGTDLAFGPALVLPLGGGESTAGVLI
Mvan_1413|M.vanbaalenii_PYR-1 GSTTGAVFQSGRPVITEAFGHSIPFFTDVGERPVVVVPLRSERQNLGVLA
MMAR_0133|M.marinum_M --------------------------------AAMVAPIVRAHRLYGLLY
MAV_5281|M.avium_104 ---------------------VVGGLLAAASAAALLLTAAALHRLDAIVL
::
Mb3156c|M.bovis_AF2122/97 VLSQGGPGAFTDEQLEMMAAFADQAALAWQLATSQRRMRELDVLTDRDRI
Rv3132c|M.tuberculosis_H37Rv VLSQGGPGAFTDEQLEMMAAFADQAALAWQLATSQRRMRELDVLTDRDRI
MUL_2422|M.ulcerans_Agy99 LLHPSSPESVTEEQLEMMAAFADQAALALQLATSQRRMRELDVVTERDRI
Mflv_3844|M.gilvum_PYR-GCK VLRSVGAQPFRSDELDMLAAFADQAAQAWQLASTQHRLRELGILTDRDRI
MSMEG_5241|M.smegmatis_MC2_155 AVQGSGARPFTAEQLEMMTGFADQAAVAWQLASSQRRMSELDILADRDRI
TH_0388|M.thermoresistible__bu VCRAEEGRPFTDEQLEILVTFADQAALAWQLAATQRRMRELDVLSDRDRI
MAB_3890c|M.abscessus_ATCC_199 LLRTVGAIAFDEEQLDMAASFAGQAALALEQAEVRLAKHELDVLADRDRI
Mvan_1413|M.vanbaalenii_PYR-1 LARNASAPRFQAGDLDLVRDYAYHTAIAIVIARARGTANELAMFTDRERI
MMAR_0133|M.marinum_M AARR-SPVAWTDRDRAELLSLARQAAVAMEVADSAREMAEVAAFGERQRL
MAV_5281|M.avium_104 YLGFIVVAWLLG------YLMRTQRLLVAEQIEAQQMLAEHAAADERRRI
: . * :* *:
Mb3156c|M.bovis_AF2122/97 ARDLHDHVIQRLFAIGLALQGAVPHERNPEVQQRLSDVVDDLQ----DVI
Rv3132c|M.tuberculosis_H37Rv ARDLHDHVIQRLFAIGLALQGAVPHERNPEVQQRLSDVVDDLQ----DVI
MUL_2422|M.ulcerans_Agy99 ARDLHDHVIQRLFAIGLALQGSVPRTREPKVQQRLSEVIDDLQ----GVI
Mflv_3844|M.gilvum_PYR-GCK ARDLHDHVIQRLFAVGLSLQGSIARARSEEVQQRLSANVDDLQ----SVI
MSMEG_5241|M.smegmatis_MC2_155 ARDLHDHVIQRLFAVGLSLQGTIPRARSAEVQQRIADCVDDLQ----AVI
TH_0388|M.thermoresistible__bu ARDLHDHVIQRLFAVGLTLQGTIPRARSTEVQQRLSECIDDLQ----AVI
MAB_3890c|M.abscessus_ATCC_199 ARDLHDHVIQRLFAVGLAMQSTHRRSVDPTVAARLADHIDQLH----EVI
Mvan_1413|M.vanbaalenii_PYR-1 AHDLHDQVIQRVFAVGMDLQGVVARVRSPALNERLSNSVDELQ----AVI
MMAR_0133|M.marinum_M AIRLHDSVGATLFSLRVALMSAQEETEPGRREELLSDALVLTE----RAT
MAV_5281|M.avium_104 AREVHDVIAHSLSITLLHVTGARRALQQDRDVDDAVEALEQAERLGRQAM
* :** : : : : . : . .
Mb3156c|M.bovis_AF2122/97 QEIRTTIYDLHGASQGITRLRQR-----IDAAVAQFA-DSGLRTSVQFVG
Rv3132c|M.tuberculosis_H37Rv QEIRTTIYDLHGASQGITRLRQR-----IDAAVAQFA-DSGLRTSVQFVG
MUL_2422|M.ulcerans_Agy99 QEIRTTIFDLHGSSQGVTRLRQR-----IDAAVAAFG-SSGLRTTVQFIG
Mflv_3844|M.gilvum_PYR-GCK QEIRTAIFDLHGGSSAVTRLRQR-----LDAAVAAFP-ESGVRTSMQCAG
MSMEG_5241|M.smegmatis_MC2_155 SEIRTAIFDLHHTAPGGTRLRQR-----LDEAIAQFS-GAGLHTTSQFVG
TH_0388|M.thermoresistible__bu GEIRTTIFDLHGGPATGSQIRQR-----IGEAANQFS-DAGVRTRVQFAG
MAB_3890c|M.abscessus_ATCC_199 QEIRTAIFDLHTNDE---SLRAT-----LRSTINELTAETGLRVSVRMSG
Mvan_1413|M.vanbaalenii_PYR-1 TEIRSTIFNLQKPAEAHGAFAKR-----IRHAFQRLTEDREMAATLTVSG
MMAR_0133|M.marinum_M SELRAQTLSLHDLPED-KALAVT-----LEGDCRDFATRTGTAAELVVLG
MAV_5281|M.avium_104 ADIRRTVGLLDNWPTKAAPTTPEPGLDDVASLVDGFR-RAGLAVTLCVEG
::* *. : : . *
Mb3156c|M.bovis_AF2122/97 PLSVVDSALADQAEAVVREAVSNAVRHAKASTLTVRVK-VDDDLCIEVTD
Rv3132c|M.tuberculosis_H37Rv PLSVVDSALADQAEAVVREAVSNAVRHAKASTLTVRVK-VDDDLCIEVTD
MUL_2422|M.ulcerans_Agy99 PLSVVDNVLADHAEAVVREAVSNAVRHAQATTLTVRVK-VDDDLCIEVSD
Mflv_3844|M.gilvum_PYR-GCK PLSVVDAGLADHAEAVVREAVSNAVRHAEASTVAVTVT-VEDDLAITVTD
MSMEG_5241|M.smegmatis_MC2_155 PLSVVDAELADHAEAVLREAVSNVVRHSGANELTVRVK-VEDDLSIEVTD
TH_0388|M.thermoresistible__bu PLSVISAVLADHAEAVVREAVSNAVRHGGATDLSVDVS-VDDDLTIEVTD
MAB_3890c|M.abscessus_ATCC_199 PLDVVPASTSGDAEAVVREAVSNVVRHARARELSVTVS-VDDELVIEVVD
Mvan_1413|M.vanbaalenii_PYR-1 PMEVVAPSLVDDAEAVVLEGLSNAVRHSGAAAVAVEIA-VGDDLVVEVCD
MMAR_0133|M.marinum_M DLPLLDSARTDALRRTAREALLNVEKHAGAHSVVVSLYRAEDGVGLTVAD
MAV_5281|M.avium_104 PADHVSPAVGLALYRITQESLANIAKHAPESKAGVVLDISPASARLAVTN
: *.: * :*. * : : * :
Mb3156c|M.bovis_AF2122/97 NG--RGLP-DEFTGSGLTNLRQRAEQAGGEFTLASVPGASGTVLRWSAPL
Rv3132c|M.tuberculosis_H37Rv NG--RGLP-DEFTGSGLTNLRQRAEQAGGEFTLASVPGASGTVLRWSAPL
MUL_2422|M.ulcerans_Agy99 NG--RGMP-KTYTSSGLTNLRRRAQEAGGQFTIESPAAGGGTLLRWSAPL
Mflv_3844|M.gilvum_PYR-GCK DG--RGIA-ADVTGSGLANLARRAQDVGGSFSVRRGAQV-GTTLTWRAPL
MSMEG_5241|M.smegmatis_MC2_155 DG--EGIS-GPVTESGLSNLRQRADQCGGELRIIDRPGG-GTILRWTAPL
TH_0388|M.thermoresistible__bu NG--RGFPEGEVTESGLANLRHRARHAGGTLEVHNRPDG-GARLHWSVPL
MAB_3890c|M.abscessus_ATCC_199 DG--IGIP-DVVARSGLSGLANRAAQSGGTFSVSAMDTG-GTRLVWSAPL
Mvan_1413|M.vanbaalenii_PYR-1 DG--QGIPAESRRRSGLANLAARARAAGGELVIDTPPSG-GTRLIWSVPL
MMAR_0133|M.marinum_M DGTAPDIASEHRRGLGLRATVERIERVGGWLSCTANEEGGGTVQAWVPAA
MAV_5281|M.avium_104 ILPAAVVAARSPEGRGLRGMRQRVELLGGAIDVGPTRDGWSVCAEIPLQE
.. ** * ** : ..
Mb3156c|M.bovis_AF2122/97 SQ-----------
Rv3132c|M.tuberculosis_H37Rv SQ-----------
MUL_2422|M.ulcerans_Agy99 IQ-----------
Mflv_3844|M.gilvum_PYR-GCK P------------
MSMEG_5241|M.smegmatis_MC2_155 PD-----------
TH_0388|M.thermoresistible__bu T------------
MAB_3890c|M.abscessus_ATCC_199 P------------
Mvan_1413|M.vanbaalenii_PYR-1 ART----------
MMAR_0133|M.marinum_M RAFRPVGP-----
MAV_5281|M.avium_104 SDGSWRPWWCGKA