For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TEEHGMNKDDLILISVDDHIAEPADMFDAHVPAKYKDRAPRVVVEPDGIQQWYYGDIRGRNMGLNAVAGK PREMYNIDASRYDEMRPGCFNVDERVRDMNAGGQLAGLNFPNFTGFSGQVLNQGPDREVNLVMIKAYNDW HIDEWCAAYPGRFIPCGILPLFDVAEAAKEVNRLAAKGCHAVTFSENPEALQMPSIHTRYWYPLFEAVCE NKTVLCTHVGSASRSPQVSTDAPPSVQMTASSMMSMFTFTELIWAQFWADFPELKFSLTEGDVGWIPYFL WRAEHVYNRHSGWTLATFPPGYSGPADVFKRHFYTCFISDKVGVRNMDWFNEDMLCWESDFPHSDSNWPF APEDVIDTMGHLDDAVINKITHENAMAAYSFDPFRHIPKEQARAGHLRAQATDVDVVTHVGHRASQRDRD AWARMTQFALQAQASAQAPVTVEAAGIASRATTLGN*
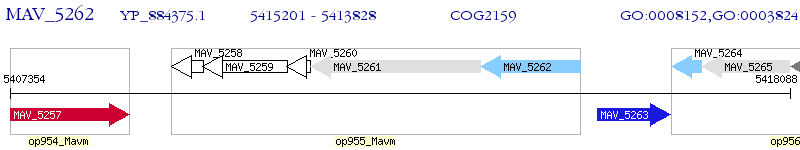
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5262 | - | - | 100% (457) | amidohydrolase 2 |
| M. avium 104 | MAV_0907 | - | e-121 | 49.07% (432) | amidohydrolase 2 |
| M. avium 104 | MAV_0860 | - | 1e-99 | 43.84% (422) | amidohydrolase 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4324 | - | 1e-111 | 46.76% (417) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4742 | - | 1e-118 | 48.58% (424) | metal-dependent amidohydrolase |
| M. smegmatis MC2 155 | MSMEG_4837 | - | 1e-118 | 49.20% (437) | amidohydrolase 2 |
| M. thermoresistible (build 8) | TH_2339 | - | 1e-101 | 42.69% (438) | amidohydrolase 2 |
| M. ulcerans Agy99 | MUL_3143 | - | 5e-72 | 38.21% (369) | amidohydrolase |
| M. vanbaalenii PYR-1 | Mvan_2022 | - | 1e-114 | 47.24% (417) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_5262|M.avium_104 MTEEHGMNKDDLILISVDDHIAEPADMFDAHVPAKYKDRAPRVVVEPDGI
TH_2339|M.thermoresistible__bu --MSHDLRPEDLILVSIDDHVVEPPDMFLRHVPAKYRDEAPIVVTDENGV
Mflv_4324|M.gilvum_PYR-GCK ------MNRDDMILISVDDHIVEPPDMFKNHLSKKYLVEAPRLVHNEDGS
Mvan_2022|M.vanbaalenii_PYR-1 ------MNRDDMILISVDDHIVEPPDMFKNHLPSKYLDEAPRLVHNPDGS
MMAR_4742|M.marinum_M ------MNKEDMILISVDDHTVEPPNMFKNHLSAKYLDDAPRLVHNPDGS
MSMEG_4837|M.smegmatis_MC2_155 ------MNKDDMILISVDDHIIEPPDMFKNHLPAKYRDEAPRLVHNPDGS
MUL_3143|M.ulcerans_Agy99 -MTDPPRARRRYTVISVDDHIVEPPDTFTGRLPQRFADRAPRVVETDDGG
::*:*** **.: * ::. :: ** :* :*
MAV_5262|M.avium_104 QQWYYGDIRGRNMGLNAVAGKPREMYNIDASRYDEMRPGCFNVDERVRDM
TH_2339|M.thermoresistible__bu DQWMYQGRPQGVSGLNAVVSWPPEEWGRNPARFAEMRPGVYDVHERVRDM
Mflv_4324|M.gilvum_PYR-GCK DTWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGCWQVDERVKDM
Mvan_2022|M.vanbaalenii_PYR-1 DTWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGCWQVDERVKDM
MMAR_4742|M.marinum_M DMWKFRDTVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGCYNVDERIKDM
MSMEG_4837|M.smegmatis_MC2_155 DTWQFRDTVIPNVALNAVAGRPKEEYGLEPQGLDEIRKGCYDPNERVKDM
MUL_3143|M.ulcerans_Agy99 QVWVYDGQVLPNVGFNAVVGRPVSEYGFEPVRFDEMRRGAWDIHERIKDM
: * : . .:***.. * . :. :. *:* * :: .**::**
MAV_5262|M.avium_104 NAGGQLAGLNFPNFT-GFSGQVLNQG-PDREVNLVMIKAYNDWHIDEWCA
TH_2339|M.thermoresistible__bu NRNGILASMCFPTFT-GFSARHLNMH-REH-VTLIMVSAYNDWHIDEWAG
Mflv_4324|M.gilvum_PYR-GCK NAGGILGSMCFPSFP-GFAGRLFAT--EDPEFSLALVQAYNDWHVEEWCG
Mvan_2022|M.vanbaalenii_PYR-1 NAGGILGSMCFPSFP-GFAGRLFAT--EDAEFSLALVQAYNDWHVEEWCG
MMAR_4742|M.marinum_M NAGGILASICFPSFP-GFAGRLFAT--EDPDFSVALVQAYNDWHIDEWCG
MSMEG_4837|M.smegmatis_MC2_155 NAGGVLATMNFPSFP-GFAARLFAT--DDEEFSLALVQAYNDWHIDEWCA
MUL_3143|M.ulcerans_Agy99 DLNGVYASLNFPSFLPGFAGQRLQQVTKDPELAWAAVRAWNDWHLEAWAG
: .* . : **.* **:.: : : . : *:****:: *..
MAV_5262|M.avium_104 AYPGRFIPCGILPLFDVAEAAKEVNRLAAKGCHAVTFSENPEALQMPSIH
TH_2339|M.thermoresistible__bu SYPGRFIPIAILPTWNPEAMCAEIRRVAAKGCRAVTMPELPHLEGLPSYH
Mflv_4324|M.gilvum_PYR-GCK AYPARFIPMTLPVIWDPEACAKEIRRNAARGVHSLTFSENPSAMGYPSFH
Mvan_2022|M.vanbaalenii_PYR-1 AYPARFIPMTLPVIWDPVACAAEIRRNAARGVHSLTFSENPSAMGYPSFH
MMAR_4742|M.marinum_M AYPARFVPMAIPVIWDAEACAAEVRRVAKKGVHALTFTENPAAMGYPSFH
MSMEG_4837|M.smegmatis_MC2_155 SNPGRFIPMAIPAIWNPGLAAAEVRRVAEKGVHSLTFTENPSTLGYPSFH
MUL_3143|M.ulcerans_Agy99 PYPERIIPCQLPWLLDPSAGAQLIHENAERGFRAVSFSENPAMLGLPSIH
. * *::* : : . :.. * :* :::::.* * ** *
MAV_5262|M.avium_104 TR-YWYPLFEAVCENKTVLCTHVGSASRSPQVSTDAPPSVQMTASSMMSM
TH_2339|M.thermoresistible__bu DEDYWGPVFRTLSEENVVMCLHIGTGFGAISMAPDAPIDNMIILATQVSA
Mflv_4324|M.gilvum_PYR-GCK DFDHWKPMWDALVDTDTVLNVHIGSSGRLAITAPDAPMDVMITLQPMNIV
Mvan_2022|M.vanbaalenii_PYR-1 DFEHWKPMWDALVDTDTVLNVHIGSSGRLAITAPDAPMDVMITLQPMNIV
MMAR_4742|M.marinum_M N-EYWNPLWKALCDTNTVMNIHIGSSGRLAITAPDAPMDVMITLQPMNIV
MSMEG_4837|M.smegmatis_MC2_155 DLEYWRPLWQALVDTETVMNVHIGSSGKLAITAPDAPMDVMITLQPMNIV
MUL_3143|M.ulcerans_Agy99 SG-HWDPMMAACAETGTVVNLHIGSSGSSPSTTDDAPADVPGVLFFAYAI
:* *: : : .*: *:*:. : *** .
MAV_5262|M.avium_104 FTFTELIWAQFWADFPELKFSLTEGDVGWIPYFLWRAEHVYNRHSGWTLA
TH_2339|M.thermoresistible__bu MCAQDLLWGPAMRNYPDLKFAFSEGGIGWIPFYLDRCDRHYTNQK-WLRR
Mflv_4324|M.gilvum_PYR-GCK QAAADLLWSRPIKEYPDLKIALSEGGTGWIPYFLERVDRTYEMHSTWTGQ
Mvan_2022|M.vanbaalenii_PYR-1 QAAADLLWSRPIKEYPDLKIALSEGGTGWIPYFLERVDRTYEMHSTWTGQ
MMAR_4742|M.marinum_M QAAADLLWSRPIKEYPDLKIALSEGGTGWIPYFLERADRTFEMHSTWTHQ
MSMEG_4837|M.smegmatis_MC2_155 QAAADLLWSAPIKEFPTLKIALSEGGTGWIPYFLDRVDRTFEMHSTWTHQ
MUL_3143|M.ulcerans_Agy99 SAAVDWLYSGLPSRFPDLKICLSEGRIGWVAGLLDRLDHMLSYHEMYGTW
: ::. :* **:.::** **:. * * :: :. :
MAV_5262|M.avium_104 TFPPGYSGPADVFKRHFYTCFISDKVGVRNMDWFNEDMLCWESDFPHSDS
TH_2339|M.thermoresistible__bu DF--GDKLPSDVFREHSLACYVTDKTSLKLRHEIGIDIIAWECDYPHSDC
Mflv_4324|M.gilvum_PYR-GCK DF--KGKLPSEVFREHFLTCFIADPVGVATRHQIGVDNICWEADYPHSDS
Mvan_2022|M.vanbaalenii_PYR-1 DF--KGKLPSEVFREHFLTCFIADPVGVATRHQIGVDNICWEADYPHSDS
MMAR_4742|M.marinum_M NF--GGKIPSEVFREHFLTCFISDKVGVALRNMIGIDNIAWEADYPHSDS
MSMEG_4837|M.smegmatis_MC2_155 NF--GGKLPSEVFREHFMTCFISDPVGVKNRHMIGIDNICWEMDYPHSDS
MUL_3143|M.ulcerans_Agy99 RAMGESLTSAEVFTRNFWFCAVEDKSSFVQHYRIGTENIMVESDYPHCDS
.::** .: * : * .. :. : : * *:**.*.
MAV_5262|M.avium_104 NWPFAPEDVIDTMGHL--DDAVINKITHENAMAAYSFDPFRHIPKEQARA
TH_2339|M.thermoresistible__bu FWPDAPEQVHAELRAAGADDFDINKITWENSCRFFSWDPFEHTPRAEATV
Mflv_4324|M.gilvum_PYR-GCK MWPGAPEQLDEVLKANNVPDDEINKMTYENAMRWYHWDPFTHIAKEQATV
Mvan_2022|M.vanbaalenii_PYR-1 MWPGAPEQLDEVLKVNNVPDDEIDKMTYQNAMRWYHWDPFTHIPKEQATV
MMAR_4742|M.marinum_M MWPGAPEELWDVLSVNDVPDGEINKMTYENAMRWYSFDPFSHITREQATV
MSMEG_4837|M.smegmatis_MC2_155 MWPGAPEELSAVFDTYDVPDDEINKITHENAMRLYHFDPFAHVPKEQATV
MUL_3143|M.ulcerans_Agy99 TWPHTQQTMHEQLDGL--PADVIRKITWENAARLYQHPVPVAIAQNPEAF
** : : : : * *:* :*: : .:
MAV_5262|M.avium_104 GHLRAQATDVDVVTHVGHRASQRDRDAWARMTQFALQAQASAQAPVTVEA
TH_2339|M.thermoresistible__bu GALRAKATDVDTT--------IRSRAEWAQLYE---QKQQFAQA------
Mflv_4324|M.gilvum_PYR-GCK GALRKAAEGHDVSIQALSHHKEGERGGGAGHFDALNAAARGNSGA-----
Mvan_2022|M.vanbaalenii_PYR-1 GALRKAAEGHDVSIQALSHHKEGERGGGAGHFDALNAAARGNSGAD----
MMAR_4742|M.marinum_M GALRKAAEGHDVSVRALSHHKDARSGSSVAEFTANAKALTGNKD------
MSMEG_4837|M.smegmatis_MC2_155 GALRKAAEGHDVSIRALS--KKDKTGASFADFQANAKAVSGARD------
MUL_3143|M.ulcerans_Agy99 --------------------------------------------------
MAV_5262|M.avium_104 AGIASRATTLGN
TH_2339|M.thermoresistible__bu ------------
Mflv_4324|M.gilvum_PYR-GCK ------------
Mvan_2022|M.vanbaalenii_PYR-1 ------------
MMAR_4742|M.marinum_M ------------
MSMEG_4837|M.smegmatis_MC2_155 ------------
MUL_3143|M.ulcerans_Agy99 ------------