For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DRRSMMLMSGIGMLGAAMRLPGAWATPPAPEAPPSAGGGPYIFADEFDGPAGSPPDPGKWTIQTWQDDVF PPVAGIYRDDRRNVFQDGNSNLVLCATQEMGTYYSGKLRGNFRSMINQTWEARIKLDCLFPGLWPSFWGV NEDPLPDGEVDIFEWYGNGQWPPGTTVHAASNGKTWEGKSIPGLVDGNWHTWRMHWGEEGFEFSRDGAEY FKVPNKPIHVAGGAPDDFRWPFNNPGYWMTPMFTLAVGGVGAGDPAAGVFPSSMLIDYIRIW*
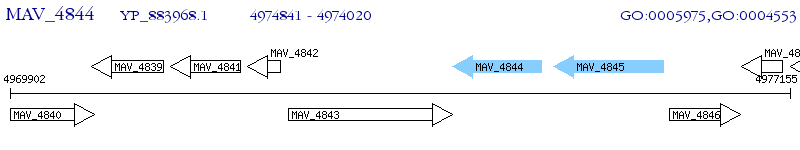
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4844 | - | - | 100% (273) | glycosyl hydrolases family protein 16 |
| M. avium 104 | MAV_2761 | - | 2e-78 | 51.41% (284) | glycosyl hydrolases family protein 16 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0323 | - | 3e-77 | 48.00% (300) | beta-1,3-glucanase precursor |
| M. gilvum PYR-GCK | Mflv_0298 | - | 7e-70 | 48.29% (292) | glycoside hydrolase family protein |
| M. tuberculosis H37Rv | Rv0315 | - | 4e-77 | 48.00% (300) | beta-1,3-glucanase precursor |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4740 | - | 7e-76 | 49.65% (288) | beta-1,3-glucanase |
| M. marinum M | MMAR_2902 | - | 4e-77 | 48.61% (288) | beta-1,3-glucanase precursor |
| M. smegmatis MC2 155 | MSMEG_5345 | - | 1e-67 | 47.54% (284) | glycosyl hydrolases family protein 16 |
| M. thermoresistible (build 8) | TH_0196 | - | 2e-69 | 45.76% (295) | POSSIBLE BETA-1,3-GLUCANASE PRECURSOR |
| M. ulcerans Agy99 | MUL_4744 | - | 1e-73 | 46.64% (298) | beta-1,3-glucanase precursor |
| M. vanbaalenii PYR-1 | Mvan_0446 | - | 3e-67 | 44.97% (298) | glycoside hydrolase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4740|M.abscessus_ATCC_1997 -----MDRRRMMAFSGLGMLAAAASMPQAWAQPSPLGPPNRPPAAPPT--
MSMEG_5345|M.smegmatis_MC2_155 --MANVDRRQMLLLSGLGATAAVLPVPRAAAVP------NQTAAS-----
MMAR_2902|M.marinum_M --MPEMDRRRMMLMTGFGVLGAALPVPAASADSSRRDMPLAPAPNGQS--
Mb0323|M.bovis_AF2122/97 MLMPEMDRRRMMMMAGFGALAAALPAPTAWADPSRPAAPAGPTPAPAAPA
Rv0315|M.tuberculosis_H37Rv MLMPEMDRRRMMMMAGFGALAAALPAPTAWADPSRPAAPAGPTPAPAAPA
MUL_4744|M.ulcerans_Agy99 MLMQEIDRRRIIMMAGFGALAAAIPAPTAGAAP---IPPAAPEPAPAAPA
TH_0196|M.thermoresistible__bu --MPMMNRRQAMMMLGFGAVAAALPLPQAGAQPVL-DAPAPGPDTP--EA
Mvan_0446|M.vanbaalenii_PYR-1 -----MDRRRAMLTLGFGMAAAALPFPKAGAQPVIGDAPPPGPSGPGLPE
Mflv_0298|M.gilvum_PYR-GCK --MPEMDRRKAMLMMGLGAAVVASPLGSAHAQ------PPAVPSEPLPAG
MAV_4844|M.avium_104 -----MDRRSMMLMSGIGMLGAAMRLPGAWATPPAPEAPPSAGGGP----
::** : *:* .. * *
MAB_4740|M.abscessus_ATCC_1997 ----GKYVFVDEFDGPAGSAPDGSKWAISKARETMKDPTFWELPENVGQY
MSMEG_5345|M.smegmatis_MC2_155 ------YVFADEFDGPAGSAPSAAKWTIAKARETIQDPTYWEQPGRIGQY
MMAR_2902|M.marinum_M ----GPYLFHDEFDGPAGSAPDSSKWTVARAREEMKDPTYWERPENVGQY
Mb0323|M.bovis_AF2122/97 AATG-GLLFHDEFDGPAGSVPDPSKWQVSNHRTPIKNPVGFDRPQFFGQY
Rv0315|M.tuberculosis_H37Rv AATG-GLLFHDEFDGPAGSVPDPSKWQVSNHRTPIKNPVGFDRPQFFGQY
MUL_4744|M.ulcerans_Agy99 AGAGPIFLWHDEFDGPAGSAPDPSKWQVSNHRTPIRNPVGFDRPEFWGQY
TH_0196|M.thermoresistible__bu ASG-PGLLFADEFDGPAGSPPDPAKWFIVPERETIRNPVEWDKPYNMGRY
Mvan_0446|M.vanbaalenii_PYR-1 AAV-PGLLFADEFNGPAGSPPNPAAWFIVPARETIRNPHEWDKPFNMGRY
Mflv_0298|M.gilvum_PYR-GCK DAG-PLMLFQDEFDGPAGAPPDPARWFIVPARETIRNPVEWDKPFNMGRY
MAV_4844|M.avium_104 ------YIFADEFDGPAGSPPDPGKWTIQTWQDDVFPPV-------AGIY
:: ***:****: *. . * : : : * * *
MAB_4740|M.abscessus_ATCC_1997 RDDRKNVFLDGNSNLVIKAAKEGNT-----YYGGKIYSTTELGIGYTWEA
MSMEG_5345|M.smegmatis_MC2_155 RNDRRNVFLDGKSNLVIRAAKDGDT-----YYSAKLASVWEGGAGHTWEA
MMAR_2902|M.marinum_M RDDRQNVFLDGKSNLVIRAAKDGGT-----YYAGKIQSPWRGGIGHTWEA
Mb0323|M.bovis_AF2122/97 RDSRQNVFLDGNSNLVLRATREGNR-----YFGGLVHGLWRGGIGTTWEA
Rv0315|M.tuberculosis_H37Rv RDSRQNVFLDGNSNLVLRATREGNR-----YFGGLVHGLWRGGIGTTWEA
MUL_4744|M.ulcerans_Agy99 RDNRQNVFLDGNSNLVLRATRSGND-----YFGGLVHGLWRGGIGTTWEA
TH_0196|M.thermoresistible__bu VTDQEHVFHDGNGNLVIRATRGPGANIREKYASAKIVGLWRGGVGTTWEA
Mvan_0446|M.vanbaalenii_PYR-1 VTDQEHVFQDGNGNLVIRATRGEGANIQEKYASAKIIGNWRGGVGTTWEA
Mflv_0298|M.gilvum_PYR-GCK VTDQEHVFQDGNGNLVIRATRGEGTTIQEKYASAKIMGTWRGGIGTTWEA
MAV_4844|M.avium_104 RDDRRNVFQDGNSNLVLCATQEMGT-----YYSGKLRGNFRSMINQTWEA
.:.:** **:.***: *:: . * .. : . . . ****
MAB_4740|M.abscessus_ATCC_1997 RIKFNCLTPGAWPAFWLGSDQ---DGEIDIVEWYGNGSWPSATTVHAKAN
MSMEG_5345|M.smegmatis_MC2_155 RIKFNCLTAGAWPAFWLGNTG---EGELDIIEWYGNGSWPSATTVHAKSN
MMAR_2902|M.marinum_M RIKFDCLTAGCWPAWWLGNQD---RGEIDIIEWYGNGSWPSATTVHAKAN
Mb0323|M.bovis_AF2122/97 RIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGTWPSGTTVHANPD
Rv0315|M.tuberculosis_H37Rv RIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGTWPSGTTVHANPD
MUL_4744|M.ulcerans_Agy99 RIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGSWPSGTTVHANPD
TH_0196|M.thermoresistible__bu RVKLNCLTDGAWPAFWLLNDDPVRGAEIDIFEWYGNRDWPSGATVHAKLD
Mvan_0446|M.vanbaalenii_PYR-1 RIKLNCLTDGAWPAFWLINDNPVRGGEVDLVEWYGNRDWPSGTTVHAKLD
Mflv_0298|M.gilvum_PYR-GCK RVKLDCLTDGAWPAFWLVNENPVRGGEVDLVEWYGNNDWPSGTTVHARLD
MAV_4844|M.avium_104 RIKLDCLFPGLWPSFWGVNEDPLPDGEVDIFEWYGNGQWPPGTTVHAASN
*:*::** * **::* . .*:*:.***** **..:**** :
MAB_4740|M.abscessus_ATCC_1997 GSEWKTHNIS--LDSGWHTWRTQWDDKGIRFWKD---YTDGATPYFEVPA
MSMEG_5345|M.smegmatis_MC2_155 GGEWKTHNIA--VDNGWHRWRTQWDAEGARFWLD---YTDGAQPYFEVAA
MMAR_2902|M.marinum_M GSEWKTRNVA--LDSGWHTWRCQWDETGMRFWQD---YAEGAQPYFTVAA
Mb0323|M.bovis_AF2122/97 GTAFETCPIG--VDGGWHNWRVTWNPSGMYFWLD---YADGIEPYFSVPA
Rv0315|M.tuberculosis_H37Rv GTAFETCPIG--VDGGWHNWRVTWNPSGMYFWLD---YADGIEPYFSVPA
MUL_4744|M.ulcerans_Agy99 GTAFETFPIG--VDDNWYNWRVTWNVTGMYFWLD---YADGMQPYFSVPA
TH_0196|M.thermoresistible__bu GTMFQTQNYP--VDSAWHTWRMTWLPSGMYFWQD---YEPGKEPFFTVLA
Mvan_0446|M.vanbaalenii_PYR-1 GTMFQTHKHP--IDSAWHTWRMTWLPTGMYFWKD---YQPGMEPFFTVAS
Mflv_0298|M.gilvum_PYR-GCK GTAFATNRHP--VDNAWHTWRMTWQPTGLYFWKD---YQPGMEPFFTVPA
MAV_4844|M.avium_104 GKTWEGKSIPGLVDGNWHTWRMHWGEEGFEFSRDGAEYFKVPNKPIHVAG
* : :*. *: ** * * * * * : * .
MAB_4740|M.abscessus_ATCC_1997 N-------SLADWPFNNPGHKAFAVLNLAVAGSGGGDPRGGTYPAEMLVD
MSMEG_5345|M.smegmatis_MC2_155 S-------SLPDWPFDQPGYTMFVVLNLAVAGSGGGDPSGGTYPADMLVD
MMAR_2902|M.marinum_M H-------SLPDWPFNDPGYTVFPVLNLAVAGSGGGDPRPGSYPAQMLVD
Mb0323|M.bovis_AF2122/97 TGIEDLNEPIREWPFNDPGYTVFPVLNLAVGGSGGGDPATGSYPQEMLVD
Rv0315|M.tuberculosis_H37Rv TGIEDLNEPIREWPFNDPGYKVFPVLNLAVGGSGGGDPATGSYPQEMLVD
MUL_4744|M.ulcerans_Agy99 TGIEDLNEPIREWPFNDPDYTVFPVLNLAVGGSGGGDPAAGTYPQEMLVD
TH_0196|M.thermoresistible__bu N-------SLPEWPFNDPGYTMVPVFNIAVGGSGGREPAGGSYPADMIID
Mvan_0446|M.vanbaalenii_PYR-1 N-------SLPDWPFNDPGYTMAPVFNIAVGGSGGREPAGGTYPAEMLVD
Mflv_0298|M.gilvum_PYR-GCK N-------SIPDWPFNDPGFTMVPVFNIAVGGSGGREPAAGTYPAEMRVD
MAV_4844|M.avium_104 G-----APDDFRWPFNNPGYWMTPMFTLAVGGVGAGDPAAGVFPSSMLID
***::*.. ::.:**.* *. :* * :* .* :*
MAB_4740|M.abscessus_ATCC_1997 WIRVW--
MSMEG_5345|M.smegmatis_MC2_155 WVRVW--
MMAR_2902|M.marinum_M WVRVW--
Mb0323|M.bovis_AF2122/97 WVRVF--
Rv0315|M.tuberculosis_H37Rv WVRVF--
MUL_4744|M.ulcerans_Agy99 WVRVF--
TH_0196|M.thermoresistible__bu WIRVF--
Mvan_0446|M.vanbaalenii_PYR-1 WIRVF--
Mflv_0298|M.gilvum_PYR-GCK WIRVFAS
MAV_4844|M.avium_104 YIRIW--
::*::