For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLMPEMDRRRMMMMAGFGALAAALPAPTAWADPSRPAAPAGPTPAPAAPAAATGGLLFHDEFDGPAGSVP DPSKWQVSNHRTPIKNPVGFDRPQFFGQYRDSRQNVFLDGNSNLVLRATREGNRYFGGLVHGLWRGGIGT TWEARIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGTWPSGTTVHANPDGTAFETCPIGVDGGWH NWRVTWNPSGMYFWLDYADGIEPYFSVPATGIEDLNEPIREWPFNDPGYKVFPVLNLAVGGSGGGDPATG SYPQEMLVDWVRVF
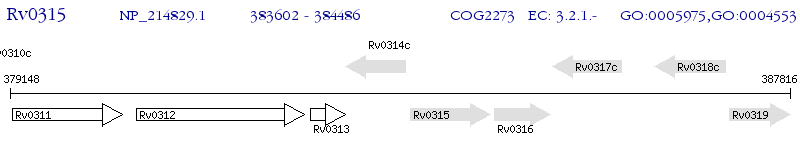
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0315 | - | - | 100% (294) | POSSIBLE BETA-1,3-GLUCANASE PRECURSOR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0323 | - | 0.0 | 99.66% (294) | beta-1,3-glucanase precursor |
| M. gilvum PYR-GCK | Mflv_0298 | - | 1e-96 | 55.22% (297) | glycoside hydrolase family protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1840c | - | 1e-106 | 62.72% (279) | putative beta-glucanase |
| M. marinum M | MMAR_0567 | - | 1e-160 | 87.12% (295) | beta-1,3-glucanase precursor |
| M. avium 104 | MAV_2761 | - | 1e-103 | 58.22% (292) | glycosyl hydrolases family protein 16 |
| M. smegmatis MC2 155 | MSMEG_0645 | - | 3e-97 | 57.79% (289) | putative beta-1,3-glucanase |
| M. thermoresistible (build 8) | TH_0196 | - | 1e-101 | 57.72% (298) | POSSIBLE BETA-1,3-GLUCANASE PRECURSOR |
| M. ulcerans Agy99 | MUL_4744 | - | 1e-159 | 86.78% (295) | beta-1,3-glucanase precursor |
| M. vanbaalenii PYR-1 | Mvan_0446 | - | 1e-96 | 56.12% (294) | glycoside hydrolase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
TH_0196|M.thermoresistible__bu --MPMMNRRQAMMMLGFGAVAAALPLPQAGAQPVL----DAPAPGPDTP-
Mvan_0446|M.vanbaalenii_PYR-1 -----MDRRRAMLTLGFGMAAAALPFPKAGAQPVIG---DAPPPGPSGPG
Mflv_0298|M.gilvum_PYR-GCK --MPEMDRRKAMLMMGLGAAVVASPLGSAHAQPPA-----VPSEP--LP-
MSMEG_0645|M.smegmatis_MC2_155 ----------MMFMMGLGVAAAALPVGTAGADPLIG---DNPPGP--GP-
Rv0315|M.tuberculosis_H37Rv MLMPEMDRRRMMMMAGFGALAAALPAPTAWADPSRPAAPAGPTPAPAAP-
Mb0323|M.bovis_AF2122/97 MLMPEMDRRRMMMMAGFGALAAALPAPTAWADPSRPAAPAGPTPAPAAP-
MMAR_0567|M.marinum_M MLMQEIDRRRMIMMAGFGALAAAIPAPTAGAAP---TPPAAPEPAPAAP-
MUL_4744|M.ulcerans_Agy99 MLMQEIDRRRIIMMAGFGALAAAIPAPTAGAAP---IPPAAPEPAPAAP-
MAV_2761|M.avium_104 --MPEIDRRRMIVTTGIGVLAAALPNPRAGASP---------APPPAAP-
MAB_1840c|M.abscessus_ATCC_199 --------------MGFGMAAAALPLPKAQADR---------VHPPAAP-
*:* ..* * * * *
TH_0196|M.thermoresistible__bu -EAASGPGLLFADEFDGPAGSPPDPAKWFIVPERETIRNPVEWDKPYNMG
Mvan_0446|M.vanbaalenii_PYR-1 LPEAAVPGLLFADEFNGPAGSPPNPAAWFIVPARETIRNPHEWDKPFNMG
Mflv_0298|M.gilvum_PYR-GCK -AGDAGPLMLFQDEFDGPAGAPPDPARWFIVPARETIRNPVEWDKPFNMG
MSMEG_0645|M.smegmatis_MC2_155 -AAAAEPTFLFQDEFNGPAGSPPDPAWWYLVPARETIKNPVEWDKPYNMG
Rv0315|M.tuberculosis_H37Rv -AAATG-GLLFHDEFDGPAGSVPDPSKWQVSNHRTPIKNPVGFDRPQFFG
Mb0323|M.bovis_AF2122/97 -AAATG-GLLFHDEFDGPAGSVPDPSKWQVSNHRTPIKNPVGFDRPQFFG
MMAR_0567|M.marinum_M -AAGAGPIFLWHDEFDGPAGSAPDPAKWQVSNHRTPIRNPVGFDRPEFWG
MUL_4744|M.ulcerans_Agy99 -AAGAGPIFLWHDEFDGPAGSAPDPSKWQVSNHRTPIRNPVGFDRPEFWG
MAV_2761|M.avium_104 -SGQSG-TYLFQDEFDGPAGSAPDASKWAVAKARETIKDPTYWERPENIG
MAB_1840c|M.abscessus_ATCC_199 -PDAQSSGFVFRDEFDGPAGSAPDGSKWVAAKFRERIKNPVFWDRPENMG
:: ***:****: *: : * * *::* :::* *
TH_0196|M.thermoresistible__bu RYVTDQEHVFHDGNGNLVIRATRGPGANIREKYASAKIVGLWRGGVGTTW
Mvan_0446|M.vanbaalenii_PYR-1 RYVTDQEHVFQDGNGNLVIRATRGEGANIQEKYASAKIIGNWRGGVGTTW
Mflv_0298|M.gilvum_PYR-GCK RYVTDQEHVFQDGNGNLVIRATRGEGTTIQEKYASAKIMGTWRGGIGTTW
MSMEG_0645|M.smegmatis_MC2_155 RYVTDQEHAFQDGKGNLVIRATRGPGTTIQEKYASAKVVGLWRGGIGTTW
Rv0315|M.tuberculosis_H37Rv QYRDSRQNVFLDGNSNLVLRATR-EGN----RYFGGLVHGLWRGGIGTTW
Mb0323|M.bovis_AF2122/97 QYRDSRQNVFLDGNSNLVLRATR-EGN----RYFGGLVHGLWRGGIGTTW
MMAR_0567|M.marinum_M QYRDNRQNVFLDGNSNLVLRATR-SGN----DYFGGLVHGLWRGGIGTTW
MUL_4744|M.ulcerans_Agy99 QYRDNRQNVFLDGNSNLVLRATR-SGN----DYFGGLVHGLWRGGIGTTW
MAV_2761|M.avium_104 QYRDDRRNVFLDGKSNLVLRAAK-DGP----TYYSGKVQSLWRGGVGHTW
MAB_1840c|M.abscessus_ATCC_199 EYRDSRTNIFLDGNSNLVIRATK-EGN----KYFSGKLHGTFRGGMGHTF
.* .: : * **:.***:**:: * * .. : . :***:* *:
TH_0196|M.thermoresistible__bu EARVKLNCLTDGAWPAFWLLNDDPVRGAEIDIFEWYGNRDWPSGATVHAK
Mvan_0446|M.vanbaalenii_PYR-1 EARIKLNCLTDGAWPAFWLINDNPVRGGEVDLVEWYGNRDWPSGTTVHAK
Mflv_0298|M.gilvum_PYR-GCK EARVKLDCLTDGAWPAFWLVNENPVRGGEVDLVEWYGNNDWPSGTTVHAR
MSMEG_0645|M.smegmatis_MC2_155 EARVKLNCLTDGAWPAFWLLNDNPVRGGEVDLVEWYGNRDWPSGTTVHAR
Rv0315|M.tuberculosis_H37Rv EARIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGTWPSGTTVHAN
Mb0323|M.bovis_AF2122/97 EARIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGTWPSGTTVHAN
MMAR_0567|M.marinum_M EARIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGSWPSGTTVHAN
MUL_4744|M.ulcerans_Agy99 EARIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGSWPSGTTVHAN
MAV_2761|M.avium_104 EARIKLDCLTAGAWPAYWLGNDD---QGEIDVMEWYGNGNWPSATTVHAK
MAB_1840c|M.abscessus_ATCC_199 EARIKFNCLTDGCWPAWWLLNDNPERGGEIDMAEWYGNRDWPSGTTVHAR
***:*::**: * ***:** *:: .*:*: ***** ***.:****.
TH_0196|M.thermoresistible__bu LDGTMFQTQNYPVDSAWHTWRMTWLPSGMYFWQDYEPGKEPFFTVLAN--
Mvan_0446|M.vanbaalenii_PYR-1 LDGTMFQTHKHPIDSAWHTWRMTWLPTGMYFWKDYQPGMEPFFTVASN--
Mflv_0298|M.gilvum_PYR-GCK LDGTAFATNRHPVDNAWHTWRMTWQPTGLYFWKDYQPGMEPFFTVPAN--
MSMEG_0645|M.smegmatis_MC2_155 LDGTSFATNPHPVDSDWHTWRMSWKPEGMYFWKDYAPGMEPFFSVPAN--
Rv0315|M.tuberculosis_H37Rv PDGTAFETCPIGVDGGWHNWRVTWNPSGMYFWLDYADGIEPYFSVPATGI
Mb0323|M.bovis_AF2122/97 PDGTAFETCPIGVDGGWHNWRVTWNPSGMYFWLDYADGIEPYFSVPATGI
MMAR_0567|M.marinum_M PDGTAFETFPIGVDDNWHNWRVTWNVTGMYFWLDYADGMQPYFSVPATGI
MUL_4744|M.ulcerans_Agy99 PDGTAFETFPIGVDDNWYNWRVTWNVTGMYFWLDYADGMQPYFSVPATGI
MAV_2761|M.avium_104 ANGGEWKTHNIAVDSAWHTWRCQWDEAGIRFWKDYVDGAQPYFDVPAS--
MAB_1840c|M.abscessus_ATCC_199 LDGTSFETLPVPIDSNWHTWRCTWTEAGLYFWMDYHDGMEPYLTVDAN--
:* : * :*. *:.** * *: ** ** * :*:: * :.
TH_0196|M.thermoresistible__bu -----SLPEWPFNDPGYTMVPVFNIAVGGSGGREPAGGSYPADMIIDWIR
Mvan_0446|M.vanbaalenii_PYR-1 -----SLPDWPFNDPGYTMAPVFNIAVGGSGGREPAGGTYPAEMLVDWIR
Mflv_0298|M.gilvum_PYR-GCK -----SIPDWPFNDPGFTMVPVFNIAVGGSGGREPAAGTYPAEMRVDWIR
MSMEG_0645|M.smegmatis_MC2_155 -----SLQDWPFNDPGYTMVPVFNIAVGGSGGREPAGGTYPAEMLVDWIR
Rv0315|M.tuberculosis_H37Rv EDLNEPIREWPFNDPGYKVFPVLNLAVGGSGGGDPATGSYPQEMLVDWVR
Mb0323|M.bovis_AF2122/97 EDLNEPIREWPFNDPGYTVFPVLNLAVGGSGGGDPATGSYPQEMLVDWVR
MMAR_0567|M.marinum_M EDLNEPIREWPFNDPDYTVFPVLNLAVGGSGGGDPAAGTYPQEMLVDWVR
MUL_4744|M.ulcerans_Agy99 EDLNEPIREWPFNDPDYTVFPVLNLAVGGSGGGDPAAGTYPQEMLVDWVR
MAV_2761|M.avium_104 -----SLPDWPFNGPGYTVFPVFNLAVAGSGGGDPGPGSYPADMLIDWIR
MAB_1840c|M.abscessus_ATCC_199 -----SLDDWPFNDPGYTLQPMLNLAVAGSGGGDPAGGSYPAEMLVDYVR
.: :****.*.:.: *::*:**.**** :*. *:** :* :*::*
TH_0196|M.thermoresistible__bu VF--
Mvan_0446|M.vanbaalenii_PYR-1 VF--
Mflv_0298|M.gilvum_PYR-GCK VFAS
MSMEG_0645|M.smegmatis_MC2_155 VFAG
Rv0315|M.tuberculosis_H37Rv VF--
Mb0323|M.bovis_AF2122/97 VF--
MMAR_0567|M.marinum_M VF--
MUL_4744|M.ulcerans_Agy99 VF--
MAV_2761|M.avium_104 VW--
MAB_1840c|M.abscessus_ATCC_199 VW--
*: