For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PEMDRRKAMLMMGLGAAVVASPLGSAHAQPPAVPSEPLPAGDAGPLMLFQDEFDGPAGAPPDPARWFIVP ARETIRNPVEWDKPFNMGRYVTDQEHVFQDGNGNLVIRATRGEGTTIQEKYASAKIMGTWRGGIGTTWEA RVKLDCLTDGAWPAFWLVNENPVRGGEVDLVEWYGNNDWPSGTTVHARLDGTAFATNRHPVDNAWHTWRM TWQPTGLYFWKDYQPGMEPFFTVPANSIPDWPFNDPGFTMVPVFNIAVGGSGGREPAAGTYPAEMRVDWI RVFAS*
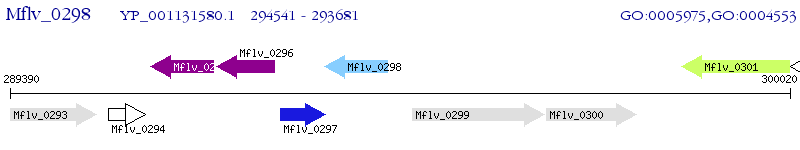
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0298 | - | - | 100% (286) | glycoside hydrolase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0323 | - | 2e-97 | 55.56% (297) | beta-1,3-glucanase precursor |
| M. tuberculosis H37Rv | Rv0315 | - | 9e-97 | 55.22% (297) | beta-1,3-glucanase precursor |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1840c | - | 2e-97 | 58.55% (275) | putative beta-glucanase |
| M. marinum M | MMAR_0567 | - | 1e-96 | 55.78% (294) | beta-1,3-glucanase precursor |
| M. avium 104 | MAV_2761 | - | 7e-93 | 52.96% (287) | glycosyl hydrolases family protein 16 |
| M. smegmatis MC2 155 | MSMEG_0645 | - | 1e-141 | 80.58% (278) | putative beta-1,3-glucanase |
| M. thermoresistible (build 8) | TH_0196 | - | 1e-137 | 74.31% (288) | POSSIBLE BETA-1,3-GLUCANASE PRECURSOR |
| M. ulcerans Agy99 | MUL_4744 | - | 2e-95 | 55.44% (294) | beta-1,3-glucanase precursor |
| M. vanbaalenii PYR-1 | Mvan_0446 | - | 1e-142 | 78.75% (287) | glycoside hydrolase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0298|M.gilvum_PYR-GCK --MPEMDRRKAMLMMGLGAAVVASPLGSAHAQPPA--VPSE----PLPAG
MSMEG_0645|M.smegmatis_MC2_155 ----------MMFMMGLGVAAAALPVGTAGADPLIGDNPPG----PGPAA
TH_0196|M.thermoresistible__bu --MPMMNRRQAMMMLGFGAVAAALPLPQAGAQPVL-DAPAPGPDTP--EA
Mvan_0446|M.vanbaalenii_PYR-1 -----MDRRRAMLTLGFGMAAAALPFPKAGAQPVIGDAPPPGPSGPGLPE
Mb0323|M.bovis_AF2122/97 MLMPEMDRRRMMMMAGFGALAAALPAPTAWADPSRPAAPAGPTPAPAAPA
Rv0315|M.tuberculosis_H37Rv MLMPEMDRRRMMMMAGFGALAAALPAPTAWADPSRPAAPAGPTPAPAAPA
MMAR_0567|M.marinum_M MLMQEIDRRRMIMMAGFGALAAAIPAPTAGAAP---TPPAAPEPAPAAPA
MUL_4744|M.ulcerans_Agy99 MLMQEIDRRRIIMMAGFGALAAAIPAPTAGAAP---IPPAAPEPAPAAPA
MAV_2761|M.avium_104 --MPEIDRRRMIVTTGIGVLAAALPNPRAGASP---------APPPAAPS
MAB_1840c|M.abscessus_ATCC_199 --------------MGFGMAAAALPLPKAQADR---------VHPPAAPP
*:* ..* * * * *
Mflv_0298|M.gilvum_PYR-GCK DAGP-LMLFQDEFDGPAGAPPDPARWFIVPARETIRNPVEWDKPFNMGRY
MSMEG_0645|M.smegmatis_MC2_155 AAEP-TFLFQDEFNGPAGSPPDPAWWYLVPARETIKNPVEWDKPYNMGRY
TH_0196|M.thermoresistible__bu ASGP-GLLFADEFDGPAGSPPDPAKWFIVPERETIRNPVEWDKPYNMGRY
Mvan_0446|M.vanbaalenii_PYR-1 AAVP-GLLFADEFNGPAGSPPNPAAWFIVPARETIRNPHEWDKPFNMGRY
Mb0323|M.bovis_AF2122/97 AATG-GLLFHDEFDGPAGSVPDPSKWQVSNHRTPIKNPVGFDRPQFFGQY
Rv0315|M.tuberculosis_H37Rv AATG-GLLFHDEFDGPAGSVPDPSKWQVSNHRTPIKNPVGFDRPQFFGQY
MMAR_0567|M.marinum_M AGAGPIFLWHDEFDGPAGSAPDPAKWQVSNHRTPIRNPVGFDRPEFWGQY
MUL_4744|M.ulcerans_Agy99 AGAGPIFLWHDEFDGPAGSAPDPSKWQVSNHRTPIRNPVGFDRPEFWGQY
MAV_2761|M.avium_104 GQSG-TYLFQDEFDGPAGSAPDASKWAVAKARETIKDPTYWERPENIGQY
MAB_1840c|M.abscessus_ATCC_199 DAQSSGFVFRDEFDGPAGSAPDGSKWVAAKFRERIKNPVFWDRPENMGEY
:: ***:****: *: : * * *::* :::* *.*
Mflv_0298|M.gilvum_PYR-GCK VTDQEHVFQDGNGNLVIRATRGEGTTIQEKYASAKIMGTWRGGIGTTWEA
MSMEG_0645|M.smegmatis_MC2_155 VTDQEHAFQDGKGNLVIRATRGPGTTIQEKYASAKVVGLWRGGIGTTWEA
TH_0196|M.thermoresistible__bu VTDQEHVFHDGNGNLVIRATRGPGANIREKYASAKIVGLWRGGVGTTWEA
Mvan_0446|M.vanbaalenii_PYR-1 VTDQEHVFQDGNGNLVIRATRGEGANIQEKYASAKIIGNWRGGVGTTWEA
Mb0323|M.bovis_AF2122/97 RDSRQNVFLDGNSNLVLRATR-EGN----RYFGGLVHGLWRGGIGTTWEA
Rv0315|M.tuberculosis_H37Rv RDSRQNVFLDGNSNLVLRATR-EGN----RYFGGLVHGLWRGGIGTTWEA
MMAR_0567|M.marinum_M RDNRQNVFLDGNSNLVLRATR-SGN----DYFGGLVHGLWRGGIGTTWEA
MUL_4744|M.ulcerans_Agy99 RDNRQNVFLDGNSNLVLRATR-SGN----DYFGGLVHGLWRGGIGTTWEA
MAV_2761|M.avium_104 RDDRRNVFLDGKSNLVLRAAK-DGP----TYYSGKVQSLWRGGVGHTWEA
MAB_1840c|M.abscessus_ATCC_199 RDSRTNIFLDGNSNLVIRATK-EGN----KYFSGKLHGTFRGGMGHTFEA
.: : * **:.***:**:: * * .. : . :***:* *:**
Mflv_0298|M.gilvum_PYR-GCK RVKLDCLTDGAWPAFWLVNENPVRGGEVDLVEWYGNNDWPSGTTVHARLD
MSMEG_0645|M.smegmatis_MC2_155 RVKLNCLTDGAWPAFWLLNDNPVRGGEVDLVEWYGNRDWPSGTTVHARLD
TH_0196|M.thermoresistible__bu RVKLNCLTDGAWPAFWLLNDDPVRGAEIDIFEWYGNRDWPSGATVHAKLD
Mvan_0446|M.vanbaalenii_PYR-1 RIKLNCLTDGAWPAFWLINDNPVRGGEVDLVEWYGNRDWPSGTTVHAKLD
Mb0323|M.bovis_AF2122/97 RIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGTWPSGTTVHANPD
Rv0315|M.tuberculosis_H37Rv RIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGTWPSGTTVHANPD
MMAR_0567|M.marinum_M RIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGSWPSGTTVHANPD
MUL_4744|M.ulcerans_Agy99 RIKFNCLAPGMWPAWWLSNDDPGRSGEIDLIEWYGNGSWPSGTTVHANPD
MAV_2761|M.avium_104 RIKLDCLTAGAWPAYWLGNDD---QGEIDVMEWYGNGNWPSATTVHAKAN
MAB_1840c|M.abscessus_ATCC_199 RIKFNCLTDGCWPAWWLLNDNPERGGEIDMAEWYGNRDWPSGTTVHARLD
*:*::**: * ***:** *:: .*:*: ***** ***.:****. :
Mflv_0298|M.gilvum_PYR-GCK GTAFATNRHPVDNAWHTWRMTWQPTGLYFWKDYQPGMEPFFTVPAN----
MSMEG_0645|M.smegmatis_MC2_155 GTSFATNPHPVDSDWHTWRMSWKPEGMYFWKDYAPGMEPFFSVPAN----
TH_0196|M.thermoresistible__bu GTMFQTQNYPVDSAWHTWRMTWLPSGMYFWQDYEPGKEPFFTVLAN----
Mvan_0446|M.vanbaalenii_PYR-1 GTMFQTHKHPIDSAWHTWRMTWLPTGMYFWKDYQPGMEPFFTVASN----
Mb0323|M.bovis_AF2122/97 GTAFETCPIGVDGGWHNWRVTWNPSGMYFWLDYADGIEPYFSVPATGIED
Rv0315|M.tuberculosis_H37Rv GTAFETCPIGVDGGWHNWRVTWNPSGMYFWLDYADGIEPYFSVPATGIED
MMAR_0567|M.marinum_M GTAFETFPIGVDDNWHNWRVTWNVTGMYFWLDYADGMQPYFSVPATGIED
MUL_4744|M.ulcerans_Agy99 GTAFETFPIGVDDNWYNWRVTWNVTGMYFWLDYADGMQPYFSVPATGIED
MAV_2761|M.avium_104 GGEWKTHNIAVDSAWHTWRCQWDEAGIRFWKDYVDGAQPYFDVPAS----
MAB_1840c|M.abscessus_ATCC_199 GTSFETLPVPIDSNWHTWRCTWTEAGLYFWMDYHDGMEPYLTVDAN----
* : * :*. *:.** * *: ** ** * :*:: * :.
Mflv_0298|M.gilvum_PYR-GCK ---SIPDWPFNDPGFTMVPVFNIAVGGSGGREPAAGTYPAEMRVDWIRVF
MSMEG_0645|M.smegmatis_MC2_155 ---SLQDWPFNDPGYTMVPVFNIAVGGSGGREPAGGTYPAEMLVDWIRVF
TH_0196|M.thermoresistible__bu ---SLPEWPFNDPGYTMVPVFNIAVGGSGGREPAGGSYPADMIIDWIRVF
Mvan_0446|M.vanbaalenii_PYR-1 ---SLPDWPFNDPGYTMAPVFNIAVGGSGGREPAGGTYPAEMLVDWIRVF
Mb0323|M.bovis_AF2122/97 LNEPIREWPFNDPGYTVFPVLNLAVGGSGGGDPATGSYPQEMLVDWVRVF
Rv0315|M.tuberculosis_H37Rv LNEPIREWPFNDPGYKVFPVLNLAVGGSGGGDPATGSYPQEMLVDWVRVF
MMAR_0567|M.marinum_M LNEPIREWPFNDPDYTVFPVLNLAVGGSGGGDPAAGTYPQEMLVDWVRVF
MUL_4744|M.ulcerans_Agy99 LNEPIREWPFNDPDYTVFPVLNLAVGGSGGGDPAAGTYPQEMLVDWVRVF
MAV_2761|M.avium_104 ---SLPDWPFNGPGYTVFPVFNLAVAGSGGGDPGPGSYPADMLIDWIRVW
MAB_1840c|M.abscessus_ATCC_199 ---SLDDWPFNDPGYTLQPMLNLAVAGSGGGDPAGGSYPAEMLVDYVRVW
.: :****.*.:.: *::*:**.**** :*. *:** :* :*::**:
Mflv_0298|M.gilvum_PYR-GCK AS
MSMEG_0645|M.smegmatis_MC2_155 AG
TH_0196|M.thermoresistible__bu --
Mvan_0446|M.vanbaalenii_PYR-1 --
Mb0323|M.bovis_AF2122/97 --
Rv0315|M.tuberculosis_H37Rv --
MMAR_0567|M.marinum_M --
MUL_4744|M.ulcerans_Agy99 --
MAV_2761|M.avium_104 --
MAB_1840c|M.abscessus_ATCC_199 --