For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AIVDRFNMKVVASAGLCGAAIALSPDAAAAPLITGGHACIQGQSGEVAPAAGGPVAGGAPAAGGAVAAGA TDMCAAPLTDMSGVPMAVPGPVPLGAPVPVPLGAPLPLGAPVPLAPPVPVVPAGAPLIALGGPAAAGAPV EAAAPIIDMSGVKDAPTGPAPTGGPVPGQPVRPGPSGAS*
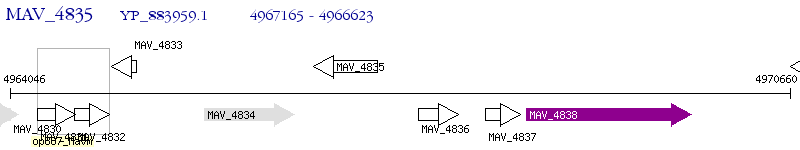
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4835 | - | - | 100% (180) | hypothetical protein MAV_4835 |
| M. avium 104 | MAV_2329 | - | 1e-10 | 36.99% (146) | hypothetical protein MAV_2329 |
| M. avium 104 | MAV_1369 | - | 4e-09 | 37.74% (106) | hypothetical protein MAV_1369 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1470c | - | 7e-56 | 57.35% (211) | proline, glycine, valine-rich secreted protein |
| M. gilvum PYR-GCK | Mflv_1398 | - | 6e-27 | 42.63% (190) | hypothetical protein Mflv_1398 |
| M. tuberculosis H37Rv | Rv1435c | - | 6e-57 | 59.31% (204) | proline, glycine, valine-rich secreted protein |
| M. leprae Br4923 | MLBr_00568 | - | 3e-21 | 38.42% (190) | hypothetical protein MLBr_00568 |
| M. abscessus ATCC 19977 | MAB_0108c | - | 5e-09 | 35.29% (119) | hypothetical protein MAB_0108c |
| M. marinum M | MMAR_2238 | - | 2e-56 | 52.02% (248) | hypothetical protein MMAR_2238 |
| M. smegmatis MC2 155 | MSMEG_0673 | - | 4e-39 | 52.38% (189) | hypothetical protein MSMEG_0673 |
| M. thermoresistible (build 8) | TH_3113 | - | 6e-07 | 31.08% (148) | PUTATIVE PPE protein |
| M. ulcerans Agy99 | MUL_1827 | - | 4e-57 | 65.95% (185) | hypothetical protein MUL_1827 |
| M. vanbaalenii PYR-1 | Mvan_2552 | - | 4e-14 | 41.88% (160) | hypothetical protein Mvan_2552 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1470c|M.bovis_AF2122/97 ------------------MTLMAIVNRFNIKVIAG-AGLFAAAIALSPDA
Rv1435c|M.tuberculosis_H37Rv ------------------MTLMAIVNRFNIKVIAG-AGLFAAAIALSPDA
MAV_4835|M.avium_104 ---------------------MAIVDRFNMKVVAS-AGLCGAAIALSPDA
MMAR_2238|M.marinum_M -MLLHQLIAEVPGANKEESALMTIFDRFNFKVVAC-AGLCGAAIALSPQA
MUL_1827|M.ulcerans_Agy99 -MLLHQLIAEVPGANKEESALMTIFDRFNFKVVAC-AGLCGAAIALSPQA
Mflv_1398|M.gilvum_PYR-GCK --------------------MMAVFDRLSIRVAAVGAGLCGIALAVSPGV
MSMEG_0673|M.smegmatis_MC2_155 ---------------------MAIFDRFNCGVVAAGAALCGVALVLSP-V
MLBr_00568|M.leprae_Br4923 ---------------------MKVFDRFNTAILVS-AGLCGAALAFSPSA
Mvan_2552|M.vanbaalenii_PYR-1 ---------------------MSRTKKSIVVVMMGSAGLFLSAPLLGPSA
MAB_0108c|M.abscessus_ATCC_199 ---------------------MADQDNSANEPNQTPEKAAGGESPKPSAP
TH_3113|M.thermoresistible__bu MALPPEVHSTLLRTGPGPGALSAAADAWRMLSDEYRAAAVELAAVVSQTC
.
Mb1470c|M.bovis_AF2122/97 AAD--PLMTGGYACIQGMAGD----------------------------A
Rv1435c|M.tuberculosis_H37Rv AAD--PLMTGGYACIQGMAGD----------------------------A
MAV_4835|M.avium_104 AAA--PLITGGHACIQGQSGE----------------------------V
MMAR_2238|M.marinum_M AAV--PLKTDGYACIQGMAGEG---------------------------G
MUL_1827|M.ulcerans_Agy99 AAV--PLKTDGYACIQGMAGEG---------------------------G
Mflv_1398|M.gilvum_PYR-GCK AHA------GGAECLQTSAGQ-----------------------------
MSMEG_0673|M.smegmatis_MC2_155 ASAVTPLVTGGDACLYEQAGT-----------------------------
MLBr_00568|M.leprae_Br4923 AAAPLSMPTGGPTCIEQMAGLG---------------------------A
Mvan_2552|M.vanbaalenii_PYR-1 NATP---TRGGVPCVGILQQVISRPGDIPQALTETARSFTNPGQPAPPVV
MAB_0108c|M.abscessus_ATCC_199 ARPAKAAKRPAAKRAPAKRPAG--------------------------SS
TH_3113|M.thermoresistible__bu ATAWEGVGADAYAAAHGPYLFWLT-------------------------D
.
Mb1470c|M.bovis_AF2122/97 PVAAGDPVAAGGPAATG-ACSAALTDMA-GVPFVAPGP------------
Rv1435c|M.tuberculosis_H37Rv PVAAGDPVAAGGPAAAG-ACSAALTDMA-GVPFVAPGP------------
MAV_4835|M.avium_104 APAAGGPVAGGAPAAGG-AVAAGATDMC-AAPLTDMSG------------
MMAR_2238|M.marinum_M APAAGGPAAAGGPAAAGGPAAAGATCAA-SAPINDMAG------------
MUL_1827|M.ulcerans_Agy99 APAAGGPAAAGGPAAAGGPAAAGATCAA-SAPINDMAG------------
Mflv_1398|M.gilvum_PYR-GCK DPAATAPC-------------APVAEMS-GVPMALPGP------------
MSMEG_0673|M.smegmatis_MC2_155 VGAAGAPAGAGAAPVGAACAAAPISEMAAGVPLAPPGP------------
MLBr_00568|M.leprae_Br4923 VPAAVPAVLPG--PVAGAVPSVPVVPSGGFPPVVPPGG------------
Mvan_2552|M.vanbaalenii_PYR-1 RPPASSSSSVGTPPVPPTPPGSVVRPSSGATPPVPPTPGTPASTTTGVGT
MAB_0108c|M.abscessus_ATCC_199 APPKSGPKAAPAPSADAAPKPPAAAKPAEAPKPAAPAP------------
TH_3113|M.thermoresistible__bu TAAAAAAAGAQLEAAAAAHASALAAMPTMAELTANRTTHAALVATNFFGV
. .
Mb1470c|M.bovis_AF2122/97 --VPAAAP------------------------------VPIGAP-VPIPG
Rv1435c|M.tuberculosis_H37Rv --VPAAAP------------------------------VPIGAP-VPIPG
MAV_4835|M.avium_104 --VPMAVPGP----------------------------VPLGAP-VPVP-
MMAR_2238|M.marinum_M --VPMAVPGP----------------------------IPVLPAGAPLID
MUL_1827|M.ulcerans_Agy99 --VPMAVPGP----------------------------IPVLPAGAPLID
Mflv_1398|M.gilvum_PYR-GCK --VPP-PMAP----------------------------PPVVPPVVPP--
MSMEG_0673|M.smegmatis_MC2_155 --VPPVPVAP----------------------------PPVVPPVVPPP-
MLBr_00568|M.leprae_Br4923 --FPPVVPPGGF--------------------------LPVVPPGGFPP-
Mvan_2552|M.vanbaalenii_PYR-1 PPVPPTPPGSTVRPAAGATPPAPPVPGVPAGTPIGGGTPPLPPVPGAPAG
MAB_0108c|M.abscessus_ATCC_199 --APAAEPAPAP--------------------------KPAAQPVSEAP-
TH_3113|M.thermoresistible__bu NTIPIAVNEADYARMWIQAATTMATYEAVASAAVSATPRTVAPPPILGAG
* .
Mb1470c|M.bovis_AF2122/97 APVPIPGAPVPIPGAPVPIP------------------------------
Rv1435c|M.tuberculosis_H37Rv APVPIPGAPVPIP-------------------------------------
MAV_4835|M.avium_104 -----LGAPLPLG-------------------------------------
MMAR_2238|M.marinum_M LGGAAAAAPVPIP-------------------------------------
MUL_1827|M.ulcerans_Agy99 LGGAAAAAPVPIP-------------------------------------
Mflv_1398|M.gilvum_PYR-GCK -----LAPP-PVVP------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 -----LAPPVPPVP------------------------------------
MLBr_00568|M.leprae_Br4923 -----VVPPGGFP-------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 TPVANVAPPSPPVPGVPVIGDGGAALPPLPPVPGGPAAVPAGVVTPPAPP
MAB_0108c|M.abscessus_ATCC_199 ------AAPKPVAP------------------------------------
TH_3113|M.thermoresistible__bu TGESATAAITGAHVTAELRAAESRTLLDEINELIDRLFAPLFPPG--FAD
.
Mb1470c|M.bovis_AF2122/97 ----GGPVPIPGAPVPVPAVPAPVIPVG----------------------
Rv1435c|M.tuberculosis_H37Rv ----GGPVPIPGAPVPVPAVPAPVIPVG----------------------
MAV_4835|M.avium_104 ------------APVPL-APPVPVVPAG----------------------
MMAR_2238|M.marinum_M ----------AAAPVPIPAGAPVPIPAGAPIPIPAGAPIPIPAGAPIPIP
MUL_1827|M.ulcerans_Agy99 ----------AAAPVPIPAG------------------------------
Mflv_1398|M.gilvum_PYR-GCK ------------PVVPPLAPP-----------------------------
MSMEG_0673|M.smegmatis_MC2_155 ------------PVVPPAGVP-----------------------------
MLBr_00568|M.leprae_Br4923 ------------PVVPPGVAPAVG--------------------------
Mvan_2552|M.vanbaalenii_PYR-1 IPGGPAAMPAGVAAPPAPAPPGAPIFGEAGAVPPPGVGGAPLDGASGVAA
MAB_0108c|M.abscessus_ATCC_199 ------------ASKPEAAKPEPAQP------------------------
TH_3113|M.thermoresistible__bu ALPDGLLGTLLAPLLPGATGDGPPMATIMSFLLMGLSNPAYLFPVLVYLL
. *
Mb1470c|M.bovis_AF2122/97 ----------------------------------TPLIALG-PVLAGAPG
Rv1435c|M.tuberculosis_H37Rv ----------------------------------TPLIALG-PVLAGAPG
MAV_4835|M.avium_104 ----------------------------------APLIALGGPAAAGAP-
MMAR_2238|M.marinum_M AGAPIPIPAGAPIPIPAGAPIPIAAGAPVPIPAGVPLIALG-PVPAGVP-
MUL_1827|M.ulcerans_Agy99 ----------------------------------VSLIALG-PVPAGVP-
Mflv_1398|M.gilvum_PYR-GCK -----------------------------------PVVP-PIPAAG----
MSMEG_0673|M.smegmatis_MC2_155 -----------------------------------PIAAAPIAAAAGPIA
MLBr_00568|M.leprae_Br4923 -----------------------------------PLLAEDSAALGSGIL
Mvan_2552|M.vanbaalenii_PYR-1 PGGPGGGAGAPGAPVAAAAGNAGPPAPGAPVGDQAGVVAPPAPPAPGAPV
MAB_0108c|M.abscessus_ATCC_199 -----------------------------------DLAAAARETAAQAKS
TH_3113|M.thermoresistible__bu FTG----------------VVHTTVAVSGMLAQLVPLMPALLPVAVAALG
: .
Mb1470c|M.bovis_AF2122/97 D-----------GVVSAPIIGMSGV--KDALTD----------------P
Rv1435c|M.tuberculosis_H37Rv D-----------GVVSAPIIGMSGV--KDALTD----------------P
MAV_4835|M.avium_104 ------------VEAAAPIIDMSGV--KDAPTG----------------P
MMAR_2238|M.marinum_M -------------AAGAPIIDMSGTG-KDGPIG----------------P
MUL_1827|M.ulcerans_Agy99 -------------AAGAPIIDMSGTG-KDGPIG----------------P
Mflv_1398|M.gilvum_PYR-GCK ------------AAGGLPVASMGGLAGKGDLTG----------------P
MSMEG_0673|M.smegmatis_MC2_155 P-----------AAAGAPLLEMSGLGGKGQPTG----------------P
MLBr_00568|M.leprae_Br4923 AGKG--------VPTASPQAALSGLVVLPGPPAS-------------VEA
Mvan_2552|M.vanbaalenii_PYR-1 GDQAGLVAPPGPPAPGVPVVDQAGVVAPPGPPAPGVPVVDEAGVVTPPGP
MAB_0108c|M.abscessus_ATCC_199 T----------VDSAPPPIPGPAPEQGRQSPNLK--------------IA
TH_3113|M.thermoresistible__bu GLAG-------VWAGLSGLAGLAGLTAVPAADGG-----------ASAAP
. .
Mb1470c|M.bovis_AF2122/97 APAGGPVP--GQPVLPGPSASAPAGAR-----------------------
Rv1435c|M.tuberculosis_H37Rv APAGGPVP--GQPVLPGPSASAPAGAR-----------------------
MAV_4835|M.avium_104 APTGGPVP--GQPVRPGPSGAS----------------------------
MMAR_2238|M.marinum_M APAGGPVA--GQPVQPGPSGAGAG--------------------------
MUL_1827|M.ulcerans_Agy99 APAGGPVA--GQPVQPGPSGAGAG--------------------------
Mflv_1398|M.gilvum_PYR-GCK APSGAPVP--GQPIPPGPTGGN----------------------------
MSMEG_0673|M.smegmatis_MC2_155 APAGAPAP--GQPAMPGPQH------------------------------
MLBr_00568|M.leprae_Br4923 PPAAAPVV--AAATPLLCCGEAAPPVR-----------------------
Mvan_2552|M.vanbaalenii_PYR-1 GAPGVPVV--GEAGVVTPPGPGAPGVPVVDEAGVVTPPGPGAPGVPVVDE
MAB_0108c|M.abscessus_ATCC_199 VGAVAAIL--ALLLLRRRRRREDSVESE----------------------
TH_3113|M.thermoresistible__bu VPAGGPSLPSGSSAPPAPTGSAAPTTGTAPGGAGSPPPAGHS--------
. . .
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 AGVVTPPGPGAPGVPVVDEAGVVTPPAPGAPGVPVDGGAELVTPPGPGAP
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu ---VPPAPHSAPPVGTSGPPYLIAAQPAPGVARRSAGARRAASIVEPAGS
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 GTPTVSSVETFSPPAPPAPGTPSSGSIETFSPPAPPTPGTPANSSVEIVT
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu AAAAAAARESAPIRRRRGQKDGLHGHRHEHQQPASMSAATAPSAAGIRVL
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 PPPPGVPGVPGEGSTTYTTPPTPEITLNPANSSIDVATPPGPGVPGVPVE
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu GGAGRISRTRGSVPHAEITPGDPTEPSSPTDRAHRPAEPN----------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 ASANVSTPPVPGVPGAPVNSSVEVATPAEVGVPGTPIERSTTLTTPPTPG
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 VPGTPVNSSVEVATPPAPGAPGVPVVDEAGVVTPPGPGAPGVPVVGEAGV
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 VTPPGPGAPGVPVVDEAGVVTPPGPGAPGVPVVGEAGVVTPPGPGAPGVP
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 VVDEAGVVTPPGPGAPGVPVVGEAGVVTPPGPGAPGVPVVDEAGVVTPPG
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 PGAPGVPVVDEAGVVTPPGPGAPGVPVVDEAGVVTPPAPPAAAPGAPIDA
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 AAASVAPSGPPAAPVLGEAGVVAPPAPPGGGAPGGGAPGAGVGTGAPIEG
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 AAGAGGVPAAPLAANAGAATPPAAATGGAGAPIDAAAGAAAPPAPPAPGA
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 PIFGEAGVAAPPLPPAPGGAGAVPAGVVTPPVPGVPGGAGAVPAGAVTPP
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 TPPVPGSSSGGFVNYPAPPVPPTPPNSAGVNTGTPGSPIPGTPGSTLTIS
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 DLLTPPGVARTVQNTASSVMSIAPPGTPPVPGGAGSVSVGVPAPPSPLTP
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 --------------------------------------------------
Rv1435c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4835|M.avium_104 --------------------------------------------------
MMAR_2238|M.marinum_M --------------------------------------------------
MUL_1827|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1398|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0673|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00568|M.leprae_Br4923 --------------------------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 LAATGLSNLVSPPSLTIPSIPGLPVPLPNEVPVPSDLLCVGTDWSATQGD
MAB_0108c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
Mb1470c|M.bovis_AF2122/97 -------------------------------
Rv1435c|M.tuberculosis_H37Rv -------------------------------
MAV_4835|M.avium_104 -------------------------------
MMAR_2238|M.marinum_M -------------------------------
MUL_1827|M.ulcerans_Agy99 -------------------------------
Mflv_1398|M.gilvum_PYR-GCK -------------------------------
MSMEG_0673|M.smegmatis_MC2_155 -------------------------------
MLBr_00568|M.leprae_Br4923 -------------------------------
Mvan_2552|M.vanbaalenii_PYR-1 SATTPVNAEIPRALVTGAAPVGDRRDRWDTR
MAB_0108c|M.abscessus_ATCC_199 -------------------------------
TH_3113|M.thermoresistible__bu -------------------------------