For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAAAVGAALVAAAFLLPRLNLGVRPRLDVGPERFATHAGAAPIFGEWLIHAGWGTGPAVALAAAVVAWGP VLAQRLSWRALTLGSWAVAAGWAFALAMIDGWQRGFAGRLTTRDEYLWEVPRVTDIAAAVRTFSSRIVDY QPHSWTTHVSGHPPGALLTFVWLDRLGLHGGGWAGLLCLLAGSSAAAAVLIGVRALADEPTARRAAPFVA LAPTAIWVAVSADGYFAGVAAWGIALLAVAVHRPLRFPALTAAGAGLLLGWGVFCNYGLMLMGLPAAAVL AAAPRPPRDGWTQWRAVLRALGPAALAAIAVAVAFAVAGFYWFDGYTLVQQRYWQGIAKDRPFQYWSWAN LACVVCAIGLGGVAGVGRVFDRAAIRRRSGFPLLLLAVLAAAACADLSMLSKAEVERIWLPFTIWLTAAA ALLPVRTHRIWLALNAVGALAVNTIILTHW
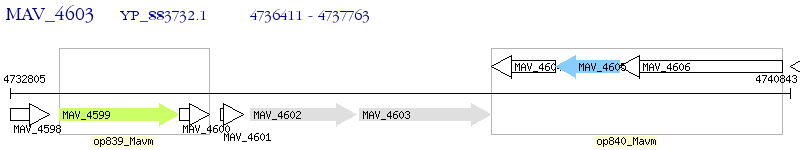
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4603 | - | - | 100% (450) | integral membrane protein |
| M. avium 104 | MAV_4843 | - | 7e-05 | 26.41% (284) | hypothetical protein MAV_4843 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0555c | - | 0.0 | 77.78% (450) | integral membrane protein |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv0541c | - | 0.0 | 77.56% (450) | integral membrane protein |
| M. leprae Br4923 | MLBr_01389 | - | 6e-05 | 26.54% (260) | hypothetical protein MLBr_01389 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0887 | - | 0.0 | 72.83% (449) | hypothetical protein MMAR_0887 |
| M. smegmatis MC2 155 | MSMEG_0999 | - | 1e-173 | 66.15% (455) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0640 | - | 0.0 | 72.16% (449) | hypothetical protein MUL_0640 |
| M. vanbaalenii PYR-1 | Mvan_0891 | - | 1e-176 | 67.41% (451) | putative integral membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0887|M.marinum_M MRIARRETAAVTVGVVLVAAAFVLPRLDLG-VKPRRDIG---LERFASRA
MUL_0640|M.ulcerans_Agy99 MRIARRETAAVTVGVVLVAAAFVLPRLDLG-VKPRRDIG---LERFASRA
Mb0555c|M.bovis_AF2122/97 MRIGRREGLAVAIGFVLVGAAFVLPRLNLG-IKPRSDIG---LERFATRA
Rv0541c|M.tuberculosis_H37Rv MRIGRREGLAVAIGFVLVGAAFVLPRLNLG-IKPRSDIG---LERFATRA
MAV_4603|M.avium_104 --------MAAAVGAALVAAAFLLPRLNLG-VRPRLDVG---PERFATHA
MSMEG_0999|M.smegmatis_MC2_155 MQIGRRELVAVIIAILLVTAAFVVPHLDLGIVTPLINST---PAQIYSFA
Mvan_0891|M.vanbaalenii_PYR-1 MPIGRKEAVAVAVGSVLVVAAFVIPHLDLGVVTPLINAT---PERLRSFA
MLBr_01389|M.leprae_Br4923 ------------MTSIMSVSALEQSAADVGDNSARQHAHGALPDSLAIAM
: : :*: . ::* . . :
MMAR_0887|M.marinum_M GAAPIFGYWDIHASWGTIAAILLALAVLVWGPRVARKLSWR-VLPLATWA
MUL_0640|M.ulcerans_Agy99 GAAPIFGYWDIHASWGTIAAILLALAVLVWGPRVARKLSWR-VLPLATWA
Mb0555c|M.bovis_AF2122/97 GAAPIFGYWDAHVGWGTAPAVLTAVAVVAWGPVVAHRLPWR-VLTLSTWA
Rv0541c|M.tuberculosis_H37Rv GAAPIFGYWDAHVGWGTAPAVLTAVAVVAWGPVVAHRLPWR-VLTLSTWA
MAV_4603|M.avium_104 GAAPIFGEWLIHAGWGTGPAVALAAAVVAWGPVLAQRLSWR-ALTLGSWA
MSMEG_0999|M.smegmatis_MC2_155 DTAPIFGWWNAHVGWGTVPAVLIGAATVWWGPAVAQRLPWR-ALPLVAWV
Mvan_0891|M.vanbaalenii_PYR-1 DTAPIFGWWNTHVGAGTIPAFAIGAAAVLWGPPLAQRLPWR-WLPWLSWT
MLBr_01389|M.leprae_Br4923 LATVISGAWASRPSLWFDEAATISASASRTVPELWRLLSHIDAVHGLYYL
:: * * * : . * . :. * : : *. : :
MMAR_0887|M.marinum_M TACGWAFSLAMIDGFERGFAGRLTTKDEYLSEVGGVTDIPVMLRTFASRI
MUL_0640|M.ulcerans_Agy99 TACGWPFSLAMIDGFERGFAGRLTTKDEYLSEVGGVTDIPVMLRTFASRI
Mb0555c|M.bovis_AF2122/97 TAAAWAFSLAMIDGWQRGFAGRLTTRDEYLWQVPGIADIPATLRTFTSRI
Rv0541c|M.tuberculosis_H37Rv TAAAWAFSLAMIDGWQRGFAGRLTTRDEYLWQVPGIADIPATLRTFTSRI
MAV_4603|M.avium_104 VAAGWAFALAMIDGWQRGFAGRLTTRDEYLWEVPRVTDIAAAVRTFSSRI
MSMEG_0999|M.smegmatis_MC2_155 TSALWAFSLAMIDGWQRGFAGRLTARHEYLRQVPSVTDIGEALRTFSSRI
Mvan_0891|M.vanbaalenii_PYR-1 VSCAWAFSLAMVDGWQRGFAGRLTARHEYLRQVPTVTDIPEALRTFAGRI
MLBr_01389|M.leprae_Br4923 LMHGWFAIFPSTEFWSRVPSCLAIGAAAAGVTVFTRQFATRTTAVYAGIV
* :. : :.* : * .::. :
MMAR_0887|M.marinum_M VDYQP-NSWTTHVSGHPPGALLTFVWLDRIGLGGGAWAG-----------
MUL_0640|M.ulcerans_Agy99 VDYQP-NSWTTHVSGHPPGALLTFVWLDRIGLGGGAWAG-----------
Mb0555c|M.bovis_AF2122/97 LDFQP-NSWVTHVSGHPPGALLTFVWLDRIGLRGGGWAG-----------
Rv0541c|M.tuberculosis_H37Rv LDFQP-NSWVTHVSGHPPGALLTFVWLDRIGLRGGGWAG-----------
MAV_4603|M.avium_104 VDYQP-HSWTTHVSGHPPGALLTFVWLDRLGLHGGGWAG-----------
MSMEG_0999|M.smegmatis_MC2_155 LDFQP-DSWITHVSGHPPGALLTFVWLDRIGLGGGAWAG-----------
Mvan_0891|M.vanbaalenii_PYR-1 LDYQP-DSWITHVSGHPPGALLTFVWLDRIGLGGGAWAG-----------
MLBr_01389|M.leprae_Br4923 FAILPRITWAGIEARSSALSVAAAMWLTVLLVASVQRNRPRLWLCYALTL
. * :* : .. :: : :** : : .
MMAR_0887|M.marinum_M FLCLLVGSSAAAAMVIAVGALAGEHVARR------------------AAP
MUL_0640|M.ulcerans_Agy99 FLCLLVGSSAAAAMVIAVGALAGKHVARR------------------AAP
Mb0555c|M.bovis_AF2122/97 LVCLLVGSSAAAAVLIAVRVLASEQMARR------------------TAP
Rv0541c|M.tuberculosis_H37Rv LVCLLVGSSAAAAVLIAVRVLASEQMARR------------------TAP
MAV_4603|M.avium_104 LLCLLAGSSAAAAVLIGVRALADEPTARR------------------AAP
MSMEG_0999|M.smegmatis_MC2_155 LLCLLVGSSAAAAIVVALRAVSGEATARL------------------AAP
Mvan_0891|M.vanbaalenii_PYR-1 LLCLLAGSSIAAAIVVTVRALADEDTARA------------------AAP
MLBr_01389|M.leprae_Br4923 MLSILLNLTLATLVLVYAVILPWLAPNKFRNSPFIWWAVTSVVALGTITP
::.:* . : *: ::: :. : :*
MMAR_0887|M.marinum_M FVAIAPTAIWIA-------------VSADGYFAGVTAWGITLLAIAVHGG
MUL_0640|M.ulcerans_Agy99 FVAIAPTAIWIA-------------VSADGYFAGVTAWGITLLAIAVHGG
Mb0555c|M.bovis_AF2122/97 FVAVAPTAIWVA-------------VSADGYFAGVAAWGIALLAVAVHGA
Rv0541c|M.tuberculosis_H37Rv FVAVAPTAIWIA-------------VSADGYFAGVAAWGIALLAVAVHGA
MAV_4603|M.avium_104 FVALAPTAIWVA-------------VSADGYFAGVAAWGIALLAVAVHRP
MSMEG_0999|M.smegmatis_MC2_155 FVAVAPTAIWIA-------------VSADGYFAGVAAWGLALLALAVRDS
Mvan_0891|M.vanbaalenii_PYR-1 FVAVAPTAIWIA-------------VSADGYFAGVAAWGITLLALAVRGS
MLBr_01389|M.leprae_Br4923 FILFAHGQVWQVDWIFRVSWHYVFDITQRQYFDHSVSFAIATAVIIVPAI
*: .* :* . :: ** .::.:: .: *
MMAR_0887|M.marinum_M SRFR---ALWAAGAGLVLGWAIFLSYGLALMGLPALAVLASANH------
MUL_0640|M.ulcerans_Agy99 SRFR---ALWAAGAGLVLGWAIFLSYGLALMGLPALAVLASANH------
Mb0555c|M.bovis_AF2122/97 TRFP---ALVAAGAGLLLGWGVFLNYGLVLIVLPGMAVLAAAD-------
Rv0541c|M.tuberculosis_H37Rv TRFP---ALVAAGAGLLLGWGVFLNYGLVLIVLPGMAVLAAAD-------
MAV_4603|M.avium_104 LRFP---ALTAAGAGLLLGWGVFCNYGLMLMGLPAAAVLAAAPRPPRDGW
MSMEG_0999|M.smegmatis_MC2_155 VHRRRVALPTAVGSGLLLGWGIFLNYGLALAALIALAVLITAPD------
Mvan_0891|M.vanbaalenii_PYR-1 VRQP---VAAAIGSGLLLGWAVFLNYGLVLMAFPAVAVLLGAAT------
MLBr_01389|M.leprae_Br4923 ATRL---AGLRAPAGDLRSLVIICTAWIVIPTTLMVGYSAVIEP------
:* : . :: . : : .
MMAR_0887|M.marinum_M --WRTALKALWPAVLAALGVALAFAVAGFWWFDGYTLVQQRYWQG---IA
MUL_0640|M.ulcerans_Agy99 --WRTALKALWPAVLAALGVALAFAVAGFWWFDGYTLVQQRYWQG---IA
Mb0555c|M.bovis_AF2122/97 --WRPVLRALGPAVLAALVVAVSFAVAGFSWFDGYTLVQQRYWQG---IA
Rv0541c|M.tuberculosis_H37Rv --WRPVLRALGPAVLAALVVAVSFAVAGFSWFDGYTLVQQRYWQG---IA
MAV_4603|M.avium_104 TQWRAVLRALGPAALAAIAVAVAFAVAGFYWFDGYTLVQQRYWQG---IA
MSMEG_0999|M.smegmatis_MC2_155 --WRAALRVAVPAVIAVLAVVAVFLVAGFWWFDGYTLVQERYWQG---IA
Mvan_0891|M.vanbaalenii_PYR-1 --WQAAVRTLVPAVVAALAVVAVFAAAGFWWFDGYLLVQERYWQG---IA
MLBr_01389|M.leprae_Br4923 VYYPRYLILTAPAAAIVIAVCIVTVARKPWPIAGVLVLFAVAAFPNYLFT
: : **. .: * . : * :: ::
MMAR_0887|M.marinum_M HDRPFQYWSWANLACVVCAIGLGSVAGISRVVDAAAIRRRSGLHLLLLG-
MUL_0640|M.ulcerans_Agy99 HDRPFQYWSWANLACVVCAIGLGSVAGISRVVDAAAIRRRSGLHLPLLG-
Mb0555c|M.bovis_AF2122/97 KDRPFGYWSWANLACVVCAIGLGSVAGLSRVFDRAAISRRSGCHLLLLA-
Rv0541c|M.tuberculosis_H37Rv KDRPFGYWSWANLACVVCAIGLGSVAGLSRVFDRAAISRRSGCHLLLLA-
MAV_4603|M.avium_104 KDRPFQYWSWANLACVVCAIGLGGVAGVGRVFDRAAIRRRSGFPLLLLA-
MSMEG_0999|M.smegmatis_MC2_155 NNRPFQYWSWANLASVVCAIGLGSVAGIGRVFDIKAARRNPGLPMLVIG-
Mvan_0891|M.vanbaalenii_PYR-1 NDRPFQYWGWANFASVVCAIGLGSVAGLGRVFDIAAIKRRSGLHLLVLG-
MLBr_01389|M.leprae_Br4923 QRGRYAKEGWD-YSQVADVISSQAAPGDCLIVDNTVPWRPGPIRALLAAR
: : .* : *. .*. ...* :.* . * : .
MMAR_0887|M.marinum_M --MVAAILVADLSMLSKAETERIWLPFTVWLTTAPA-----LLRPRSDRL
MUL_0640|M.ulcerans_Agy99 --MVAAILVADLSMLSKAETERIWLPFTVWLTTAPA-----LLRPRSDRL
Mb0555c|M.bovis_AF2122/97 --VLAAIALADLSMLSKAETERIWLPFTIWLTAAPA-----LLPPRSHRL
Rv0541c|M.tuberculosis_H37Rv --VLAAIALADLSMLSKAETERIWLPFTIWLTAAPA-----LLPPRSHRL
MAV_4603|M.avium_104 --VLAAAACADLSMLSKAEVERIWLPFTIWLTAAAA-----LLPVRTHRI
MSMEG_0999|M.smegmatis_MC2_155 --ALLAITFADLSMLSKAETERIWLPFTVWLTAAGA-----LLPPRSHRW
Mvan_0891|M.vanbaalenii_PYR-1 --ALLAIVVADLSRLSKAETERIWLPFTVWLVASTA-----LLPPRSARW
MLBr_01389|M.leprae_Br4923 PAAFRSLIDIERGFYGPTVGTLWDGHVPVWLVTAKINKCSTVWTVSDRDT
. : : . . : ..:**.:: :
MMAR_0887|M.marinum_M WLAVNAAGALAVNSIIFTNW-----------------------------
MUL_0640|M.ulcerans_Agy99 WLAVNAAGALAVNSIIFTNW-----------------------------
Mb0555c|M.bovis_AF2122/97 WLAVNAAGALLLNSIIFTNW-----------------------------
Rv0541c|M.tuberculosis_H37Rv WLAVNAAGALLLNSIIFTNW-----------------------------
MAV_4603|M.avium_104 WLALNAVGALAVNTIILTHW-----------------------------
MSMEG_0999|M.smegmatis_MC2_155 WLGLNVAGALALNHLILTNW-----------------------------
Mvan_0891|M.vanbaalenii_PYR-1 WLALNVIGALAINHLILTNW-----------------------------
MLBr_01389|M.leprae_Br4923 SLPDHQAGQLLSPGLILGRAPAYQFPSYLGFRIVERWQFHYSQVIKSTR
* : * * :*: .