For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VIVGSGFTGFTCARRLAKTLRRAPVDITMISPVDYMLYTPLLPDVAGGVVDARFVAVPLANSLRGMHAVR GRVDAVDFTGRTLTYRDPEERSHSMSWDRLVLTPGSVTKLFDVPGLARHARGLKTTAEALYLRDHFVEQL ELANIEDDPRVAAARRTVVVVGASYSGTELVAQLRALADAAARQMGFDPGAVRFLLLDLAEQVMPEVGKK LGDAALKVLRRRGIDVRLGTTLKEAHADHVVLSDGSRIDTHTIAWVTGVTGAPLIQDLGLPTEKGRLKVQ PDLQVPGHPGVFAAGDAAAVPDLTQPGKITPPTAQHATRQGKVLARNVAASLGHGSAKEYKHRNLGLVVD LGPRYAVANPLNIHLSGLPAKAVTRGYHLYAIPRGVNRWAVSLAYLTDALFARSVVSIGLSSEDDAQFTA SEGIPMPKTG
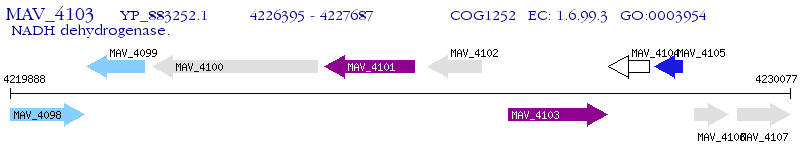
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4103 | - | - | 100% (430) | dehydrogenase |
| M. avium 104 | MAV_2867 | - | 2e-40 | 31.17% (385) | DoxD family protein/pyridine nucleotide-disulfide |
| M. avium 104 | MAV_4772 | - | 1e-38 | 30.72% (459) | DoxD family protein/pyridine nucleotide-disulfide |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1885c | ndh | 2e-38 | 29.80% (396) | NADH dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3348 | - | 4e-40 | 32.27% (375) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. tuberculosis H37Rv | Rv1854c | ndh | 1e-38 | 29.80% (396) | NADH dehydrogenase |
| M. leprae Br4923 | MLBr_02061 | - | 5e-38 | 29.04% (396) | putative dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2429c | - | 3e-41 | 31.34% (418) | NADH dehydrogenase (NDH) |
| M. marinum M | MMAR_2728 | ndh | 6e-38 | 30.23% (387) | NADH dehydrogenase Ndh |
| M. smegmatis MC2 155 | MSMEG_4433 | - | 1e-156 | 64.39% (424) | dehydrogenase |
| M. thermoresistible (build 8) | TH_2218 | - | 0.0 | 77.78% (432) | dehydrogenase |
| M. ulcerans Agy99 | MUL_3025 | ndh | 4e-38 | 30.23% (387) | NADH dehydrogenase Ndh |
| M. vanbaalenii PYR-1 | Mvan_3078 | - | 3e-39 | 31.78% (387) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2728|M.marinum_M MSPQPETTAQAR---------------PRHRVVIIGSGFGGLNAAKKLKR
MUL_3025|M.ulcerans_Agy99 MSPQPETTAQAR---------------PRHRVVIIGSGFGGLNAAKKLKR
Mb1885c|M.bovis_AF2122/97 MSPQQEPTAQPP---------------RRHRVVIIGSGFGGLNAAKKLKR
Rv1854c|M.tuberculosis_H37Rv MSPQQEPTAQPP---------------RRHRVVIIGSGFGGLNAAKKLKR
MLBr_02061|M.leprae_Br4923 MNAQPAATAAQD---------------RRHQVVIIGSGFGGLNAAKKLKR
Mflv_3348|M.gilvum_PYR-GCK MS---HPGATPS---------------DRHKVVIIGSGFGGLTAAKALKK
Mvan_3078|M.vanbaalenii_PYR-1 MS---HPGATPS---------------DRHKVVIIGSGFGGLTAAKTLKR
MAB_2429c|M.abscessus_ATCC_199 MSTTPDAAGAAAPVAANPQPKIQDALTPKHRVVIIGSGFGGLTAAKTLKR
MAV_4103|M.avium_104 --------------------------------MIVGSGFTGFTCARRLAK
TH_2218|M.thermoresistible__bu ----------------------------MTSVVIVGSGFAGFECARHLSD
MSMEG_4433|M.smegmatis_MC2_155 --------------------------MRPARIVIVGSGFAGFECARRLSK
:*:**** *: .*: *
MMAR_2728|M.marinum_M ------ADVDIKLIARTTHHLFQPLLYQVATGIISEGDIAPPTRVVLRRQ
MUL_3025|M.ulcerans_Agy99 ------ADVDIKLIARTTHHLFQPLLYQVATGIISEGDIAPPTRVVLRRQ
Mb1885c|M.bovis_AF2122/97 ------ADVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQ
Rv1854c|M.tuberculosis_H37Rv ------ADVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQ
MLBr_02061|M.leprae_Br4923 ------ANVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQ
Mflv_3348|M.gilvum_PYR-GCK ------ADVDIKMIARTTHHLFQPLLYQVATGIISEGEIAPATRVILRNQ
Mvan_3078|M.vanbaalenii_PYR-1 ------ADVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPATRVILRNQ
MAB_2429c|M.abscessus_ATCC_199 ------ANADVTLIARTTHHLFQPLLYQVATGIISEGEIAPPTRQILKDQ
MAV_4103|M.avium_104 TLRR--APVDITMISPVDYMLYTPLLPDVAGGVVDARFVAVPLANSLR--
TH_2218|M.thermoresistible__bu KLRRQQAPVDITMISPVDYMLYTPLLPDVAGGVVDARFVTVPLANTLR--
MSMEG_4433|M.smegmatis_MC2_155 QLKRHRSEAVVTLISPVDYLLYTPLLPEVAGGATDPRFATVPLNDRLP--
: . :.:*: . : *: *** :** * . : . *
MMAR_2728|M.marinum_M RNAQVLLGNVTHIDLAKQSVVS-DLLGHTYETPYDTLIVAAGAGQSYFGN
MUL_3025|M.ulcerans_Agy99 RNAQVLLGNVTHIDLAKQSVVS-DLLGHTYETPYDTLIVAAGAGQSYFGN
Mb1885c|M.bovis_AF2122/97 RNVQVLLGNVTHIDLAGQCVVS-ELLGHTYQTPYDSLIVAAGAGQSYFGN
Rv1854c|M.tuberculosis_H37Rv RNVQVLLGNVTHIDLAGQCVVS-ELLGHTYQTPYDSLIVAAGAGQSYFGN
MLBr_02061|M.leprae_Br4923 RNIQVLLGNVTHIDLANQCVVS-ELLGHTYETLYDSLIVAAGAGQSYFGN
Mflv_3348|M.gilvum_PYR-GCK KNAQVLLGDVTRIDLKTNTVRS-ELLGHTYITPFDSLIVAAGAGQSYFGN
Mvan_3078|M.vanbaalenii_PYR-1 KNAQVLLGDVTGIDLQTKTVRS-ELLGHTYVTPFDSLIVAAGAGQSYFGN
MAB_2429c|M.abscessus_ATCC_199 DNAQVVLGDVTSIDLEKQVVHS-SLLGHDYSTPYDSLIVAAGAGQSYFGN
MAV_4103|M.avium_104 -GMHAVRGRVDAVDFTGRTLTYRDPEERSHSMSWDRLVLTPGSVTKLFDV
TH_2218|M.thermoresistible__bu -GVRTARGWVDSVDFDARTVSFTDPEDRTHTVAWDRLVLTPGSVTRLFDI
MSMEG_4433|M.smegmatis_MC2_155 -EVQFIRAYVDSVDLSARSLRCRDQENMSHELSWDRLVLTPGSISRLFDI
: . * :*: . : . : :* *:::.*: *.
MMAR_2728|M.marinum_M DHFAEFAPGMKSIDDALELRGRILSAFEQAERSRDAERRAKLLTFTVVGA
MUL_3025|M.ulcerans_Agy99 DHFAEFAPGMKSIDDALELRGRILSAFEQAERSRDAERRAKLLTFTVVGA
Mb1885c|M.bovis_AF2122/97 DHFAEFAPGMKSIDDALELRGRILSAFEQAERSSDPERRAKLLTFTVVGA
Rv1854c|M.tuberculosis_H37Rv DHFAEFAPGMKSIDDALELRGRILSAFEQAERSSDPERRAKLLTFTVVGA
MLBr_02061|M.leprae_Br4923 DQFAEFAPGMKSIDDALELRGRILSAFEQAERSNDPERREKLLTFTVVGA
Mflv_3348|M.gilvum_PYR-GCK DHFAEWAPGMKSIDDALELRGRILGSFEQAERSSDPARREKLLTFVVVGA
Mvan_3078|M.vanbaalenii_PYR-1 DHFAEWAPGMKSIDDALELRGRILGAFEQAERSSDPARREKLLTFVVVGA
MAB_2429c|M.abscessus_ATCC_199 DHFAEWAPGMKSIDDALELRGRILGAFEQAERSSDPVRRRKLMTFTVVGA
MAV_4103|M.avium_104 PGLARHARGLKTTAEALYLRDHFVEQLELANIEDDPRVAAARRTVVVVGA
TH_2218|M.thermoresistible__bu PGLAEHARGLKSTAEALYLRDHFLQQLQLSCIDDDPDEIAARRTVVVVGA
MSMEG_4433|M.smegmatis_MC2_155 PGLAEHGRGLKSTAQALFLRDHIIEQFELARIDEDRERANARKTVVVVGS
:*. . *:*: :** **.::: :: : . * *..***:
MMAR_2728|M.marinum_M GPTGVEMAGQIAELAEHTLKGAFRHIDSTRARVILLDAAPAVLPPFGDKL
MUL_3025|M.ulcerans_Agy99 GPTGVEMAGQIAELAEHTLKGAFRHIDSTRARVILLDAAPAVLPPFGDKL
Mb1885c|M.bovis_AF2122/97 GPTGVEMAGQIAELAEHTLKGAFRHIDSTKARVILLDAAPAVLPPMGAKL
Rv1854c|M.tuberculosis_H37Rv GPTGVEMAGQIAELAEHTLKGAFRHIDSTKARVILLDAAPAVLPPMGAKL
MLBr_02061|M.leprae_Br4923 GPTGVEMAGQIAELADHTLKGAFRRIDSTKARVVLLDAAPAVLPPMGEKL
Mflv_3348|M.gilvum_PYR-GCK GPTGVEMAGQIAELADHTLKGAFRHIDSTKARVILLDAAPAVLPPMGEKL
Mvan_3078|M.vanbaalenii_PYR-1 GPTGVEMAGQIAELADHTLKGAFRHIDSTKARVILLDAAPAVLPPMGEKL
MAB_2429c|M.abscessus_ATCC_199 GPTGVEMAGQIAELANETLKGTFRHIDPTDARVILLDAAPAVLPPFGPKL
MAV_4103|M.avium_104 SYSGTELVAQLRALADAAARQMG--FDPGAVRFLLLDLAEQVMPEVGKKL
TH_2218|M.thermoresistible__bu SYSGTELVAQLRALADTAAEQMG--FDPAEVRFVLLDLAEQVMPEVGEKL
MSMEG_4433|M.smegmatis_MC2_155 SYSGTELIAQLCAMASVAVHDFG--FEPGDVRLILVERSGRLMPEIGPAL
. :*.*: .*: :*. : . ::. .*.:*:: : ::* .* *
MMAR_2728|M.marinum_M GERAAARLQKLGVEIQLGAMVTDVDRNGITVKDSDGTIRRIESACKVWSA
MUL_3025|M.ulcerans_Agy99 GERAAARLQKLGVEIQLGAMVTDVDRNGITVKDSDGTIRRIESACKVWSA
Mb1885c|M.bovis_AF2122/97 GQRAAARLQKLGVEIQLGAMVTDVDRNGITVKDSDGTVRRIESACKVWSA
Rv1854c|M.tuberculosis_H37Rv GQRAAARLQKLGVEIQLGAMVTDVDRNGITVKDSDGTVRRIESACKVWSA
MLBr_02061|M.leprae_Br4923 GQRAAARLQKMGVEIQLSAMVTDVDRNGITVQDSDGTVRRIESACKVWSA
Mflv_3348|M.gilvum_PYR-GCK GKQAQKRLEKMGVEIQLNAMVTDVDRNGITVKDPDGTMRRIEAACKVWSA
Mvan_3078|M.vanbaalenii_PYR-1 GKKAQARLEKLGVEIQLNAMVTDVDRNGITVKDPDGTIRRIEAACKVWSA
MAB_2429c|M.abscessus_ATCC_199 GDKARKRLEKLGVEIQLSAMVTDVDRNGLTVKHADGSIERIESWCKVWSA
MAV_4103|M.avium_104 GDAALKVLRRRGIDVRLGTTLKEAHADHVVLSDGS----RIDTHTIAWVT
TH_2218|M.thermoresistible__bu GTAAMQVLRHRGIDVRLGLTLSEVHADHVVLSDGS----RVGTRTVAWLT
MSMEG_4433|M.smegmatis_MC2_155 AVRAEHVLKRRGVRFLTGVTLSRVAADHVILSDGT----RIGTYTVAWVA
. * *.: *: . . :. . : : :.. *: : .* :
MMAR_2728|M.marinum_M GVQASRLGRDLAEQSEVELDRAGRVQVLPDLSVPGHPNVFVIGDMAAVEG
MUL_3025|M.ulcerans_Agy99 GVQASRLGRDLAEQSEVELDRAGRVQVLPDLSVPGHPNVFVIGDMAAVEG
Mb1885c|M.bovis_AF2122/97 GVSASWLGRDLAEQSRVELDRAGRVQVLPDLSIPGYPNVFVVGDMAAVEG
Rv1854c|M.tuberculosis_H37Rv GVSASRLGRDLAEQSRVELDRAGRVQVLPDLSIPGYPNVFVVGDMAAVEG
MLBr_02061|M.leprae_Br4923 GVSASRLGRDLAEQSLVELDWAGRVKVLPDLSIPGYRNVFVVGDMAAIDG
Mflv_3348|M.gilvum_PYR-GCK GVSASPLGRDLADQSGVELDRAGRVKVLPDLSIPGHPNVFVVGDMAAVED
Mvan_3078|M.vanbaalenii_PYR-1 GVSASPLGRDLAEGSGVELDRAGRVKVLPDLSLPGHPNVFVVGDMAAVEG
MAB_2429c|M.abscessus_ATCC_199 GVSASPLGKNLAEQSGVELDRAGRVKVGPDLSIPGHPNVFVVGDMMAVDG
MAV_4103|M.avium_104 GVTGAPLIQDLG-----LPTEKGRLKVQPDLQVPGHPGVFAAGDAAAVPD
TH_2218|M.thermoresistible__bu GVTAAPLIETLG-----LPTTKGRLEVDAELRVPDRPDVFAAGDAAAVPD
MSMEG_4433|M.smegmatis_MC2_155 GVVASPLITATG-----LPTEKGRLKVGPDLQVPGFDGVFAGGDAAAVPD
** .: * . **::* .:* :*. .**. ** *: .
MMAR_2728|M.marinum_M ---------VPGVAQGAIQGAKYVATTIKSELAGANAAEREPFQYFDKGS
MUL_3025|M.ulcerans_Agy99 ---------VPGVAQGAIQGAKYVATTIKSELAGANAAEREPFQYFDKGS
Mb1885c|M.bovis_AF2122/97 ---------VPGVAQGAIQGAKYVASTIKAELAGANPAEREPFQYFDKGS
Rv1854c|M.tuberculosis_H37Rv ---------VPGVAQGAIQGAKYVASTIKAELAGANPAEREPFQYFDKGS
MLBr_02061|M.leprae_Br4923 ---------VPGVAQGAIQGAKYVANNIKAELGEANPAEREPFQYFDKGS
Mflv_3348|M.gilvum_PYR-GCK ---------VPGMAQGAIQGGRYAAKAIAAGLKGAEVSEREPFKYFDKGS
Mvan_3078|M.vanbaalenii_PYR-1 ---------VPGMAQGAIQGGKYAAKAIAAGLKGADPTEREPFKYFDKGS
MAB_2429c|M.abscessus_ATCC_199 ---------VPGVAQGAIQGGRYAAKQIKAELKGRSPAERAPFKFFDKGS
MAV_4103|M.avium_104 LTQPG--KITPPTAQHATRQGKVLARNVAASLG---HGSAKEYKHRNLGL
TH_2218|M.thermoresistible__bu LAQPDPERITPPTAQHATRQGKVLANNVAASLG---YGRMKQYRHRNMGL
MSMEG_4433|M.smegmatis_MC2_155 VTNPG--TITPPTAQHATRQGKILARNVAASLG---YGSAREYRHRDLGT
.* ** * : .: * : : * ::. : *
MMAR_2728|M.marinum_M MATVSRFSAVAKIGPVEISGFIAWLAWLVLHLVYLVGFKTKVSTLLSWTV
MUL_3025|M.ulcerans_Agy99 MATVSRFSAVAKIGPVEISGFIAWLAWLVLHLVYLVGFKTKVSTLLSWTV
Mb1885c|M.bovis_AF2122/97 MATVSRFSAVAKIGPVEFSGFIAWLIWLVLHLAYLIGFKTKITTLLSWTV
Rv1854c|M.tuberculosis_H37Rv MATVSRFSAVAKIGPVEFSGFIAWLIWLVLHLAYLIGFKTKITTLLSWTV
MLBr_02061|M.leprae_Br4923 MATVSRFSAVAKIGPLEFSGFIAWLMWLVLHLMYLIGFKTKITTLLSWTV
Mflv_3348|M.gilvum_PYR-GCK MATVSRFSAVAKIGPLEFGGFIAWLSWLVLHLVYLVGFKTKIVTLLSWTV
Mvan_3078|M.vanbaalenii_PYR-1 MATVSRFSAVAKIGPLEFEGFVAWLAWLVLHLVYLVGFKTKIVTLLSWTV
MAB_2429c|M.abscessus_ATCC_199 MATVSRYSAVVKIGKIEFSGFIAWLSWLALHLVYLVGFKNRLVTLILWSI
MAV_4103|M.avium_104 VVDLGPRYAVANPLNIHLSGLPAKAVTRGYHLYAIPRGVNRWAVSLAYLT
TH_2218|M.thermoresistible__bu VVDLGPRYAVANPLNVHLSGFPAKAVARGYHLYAIPRFVNRWAVALAYLT
MSMEG_4433|M.smegmatis_MC2_155 VVDLGPGFAVANPLNIPLSGFLAKLVTRAYHLYAIPRNANRWAVALGYLT
:. :. **.: : : *: * ** : .: . : :
MMAR_2728|M.marinum_M TFLSTRRGQLTITEQQAFARTRLEQLAELASEAQ--------STEARQAS
MUL_3025|M.ulcerans_Agy99 TFLSTRRGQLTITEQQAFARTRLEQLAELASEAQ--------STEARQAS
Mb1885c|M.bovis_AF2122/97 TFLSTRRGQLTITDQQAFARTRLEQLAELAAEAQG------SAASAKVAS
Rv1854c|M.tuberculosis_H37Rv TFLSTRRGQLTITDQQAFARTRLEQLAELAAEAQG------SAASAKVAS
MLBr_02061|M.leprae_Br4923 TFLSTRRGQLTITEQQAFARTRLEQLAELTAESQS------TAAANRATM
Mflv_3348|M.gilvum_PYR-GCK TFLSTKRGQLTITEQQAYARIRLEELEEIAASVQ---------DTEKAAS
Mvan_3078|M.vanbaalenii_PYR-1 TFLSTKRGQLTITEQQAYARTRIEELEEIAASVQ---------ETERAAS
MAB_2429c|M.abscessus_ATCC_199 AFVTRSRGQMAITEQQAWARTRLDQLEELVAEKNGDEEPTTQPDKEKAAS
MAV_4103|M.avium_104 DALFARSVVSIGLSSEDDAQFTASEG-IPMPKTG----------------
TH_2218|M.thermoresistible__bu DALFDRSVVSIGLSSKEDASFAVSQR-IPPPKQPTPGD------------
MSMEG_4433|M.smegmatis_MC2_155 NVLFGRPLVSIGFAQPSWAQFSASEEFMTAPTATQPLDEQMYTDDLRFQE
: . * .: .
MMAR_2728|M.marinum_M -----------------
MUL_3025|M.ulcerans_Agy99 -----------------
Mb1885c|M.bovis_AF2122/97 -----------------
Rv1854c|M.tuberculosis_H37Rv -----------------
MLBr_02061|M.leprae_Br4923 RAS--------------
Mflv_3348|M.gilvum_PYR-GCK -----------------
Mvan_3078|M.vanbaalenii_PYR-1 -----------------
MAB_2429c|M.abscessus_ATCC_199 -----------------
MAV_4103|M.avium_104 -----------------
TH_2218|M.thermoresistible__bu -----------------
MSMEG_4433|M.smegmatis_MC2_155 ASADRASAPGDRARKAT