For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSPQPETTAQARPRHRVVIIGSGFGGLNAAKKLKRADVDIKLIARTTHHLFQPLLYQVATGIISEGDIAP PTRVVLRRQRNAQVLLGNVTHIDLAKQSVVSDLLGHTYETPYDTLIVAAGAGQSYFGNDHFAEFAPGMKS IDDALELRGRILSAFEQAERSRDAERRAKLLTFTVVGAGPTGVEMAGQIAELAEHTLKGAFRHIDSTRAR VILLDAAPAVLPPFGDKLGERAAARLQKLGVEIQLGAMVTDVDRNGITVKDSDGTIRRIESACKVWSAGV QASRLGRDLAEQSEVELDRAGRVQVLPDLSVPGHPNVFVIGDMAAVEGVPGVAQGAIQGAKYVATTIKSE LAGANAAEREPFQYFDKGSMATVSRFSAVAKIGPVEISGFIAWLAWLVLHLVYLVGFKTKVSTLLSWTVT FLSTRRGQLTITEQQAFARTRLEQLAELASEAQSTEARQAS
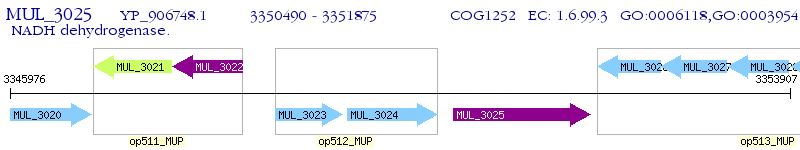
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3025 | ndh | - | 100% (461) | NADH dehydrogenase Ndh |
| M. ulcerans Agy99 | MUL_3068 | - | 1e-20 | 31.08% (325) | NADH dehydrogenase |
| M. ulcerans Agy99 | MUL_4658 | - | 2e-16 | 28.16% (348) | dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1885c | ndh | 0.0 | 91.25% (457) | NADH dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3348 | - | 0.0 | 81.72% (443) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. tuberculosis H37Rv | Rv1854c | ndh | 0.0 | 91.47% (457) | NADH dehydrogenase |
| M. leprae Br4923 | MLBr_02061 | - | 0.0 | 86.87% (457) | putative dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2429c | - | 0.0 | 75.60% (455) | NADH dehydrogenase (NDH) |
| M. marinum M | MMAR_2728 | ndh | 0.0 | 100.00% (461) | NADH dehydrogenase Ndh |
| M. avium 104 | MAV_2867 | - | 0.0 | 88.29% (461) | DoxD family protein/pyridine nucleotide-disulfide |
| M. smegmatis MC2 155 | MSMEG_3621 | ndh | 0.0 | 81.70% (448) | NADH dehydrogenase |
| M. thermoresistible (build 8) | TH_1173 | ndh | 0.0 | 81.76% (444) | PROBABLE NADH DEHYDROGENASE NDH |
| M. vanbaalenii PYR-1 | Mvan_3078 | - | 0.0 | 83.52% (443) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3348|M.gilvum_PYR-GCK ------------------MSHPGATPSDRHKVVIIGSGFGGLTAAKALKK
Mvan_3078|M.vanbaalenii_PYR-1 ------------------MSHPGATPSDRHKVVIIGSGFGGLTAAKTLKR
MSMEG_3621|M.smegmatis_MC2_155 ------------------MSHPGATASDRHKVVIIGSGFGGLTAAKTLKR
TH_1173|M.thermoresistible__bu -------------------------MSDRHKVVIIGSGFGGLNAAKALRR
Mb1885c|M.bovis_AF2122/97 ---------------MSPQQEPTAQPPRRHRVVIIGSGFGGLNAAKKLKR
Rv1854c|M.tuberculosis_H37Rv ---------------MSPQQEPTAQPPRRHRVVIIGSGFGGLNAAKKLKR
MLBr_02061|M.leprae_Br4923 ---------------MNAQPAATAAQDRRHQVVIIGSGFGGLNAAKKLKR
MUL_3025|M.ulcerans_Agy99 ---------------MSPQPETTAQARPRHRVVIIGSGFGGLNAAKKLKR
MMAR_2728|M.marinum_M ---------------MSPQPETTAQARPRHRVVIIGSGFGGLNAAKKLKR
MAV_2867|M.avium_104 ---------------MSPHSGSTAGPERRHQVVIIGSGFGGLNAAKKLKH
MAB_2429c|M.abscessus_ATCC_199 MSTTPDAAGAAAPVAANPQPKIQDALTPKHRVVIIGSGFGGLTAAKTLKR
:*:***********.*** *::
Mflv_3348|M.gilvum_PYR-GCK ADVDIKMIARTTHHLFQPLLYQVATGIISEGEIAPATRVILRNQKNAQVL
Mvan_3078|M.vanbaalenii_PYR-1 ADVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPATRVILRNQKNAQVL
MSMEG_3621|M.smegmatis_MC2_155 ADVDVKLIARTTHHLFQPLLYQVATGIISEGEIAPATRVILRKQKNAQVL
TH_1173|M.thermoresistible__bu ADVDVKLIARTTHHLFQPLLYQVATGIISEGEIAPATRVVLRRQRNVQVL
Mb1885c|M.bovis_AF2122/97 ADVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQRNVQVL
Rv1854c|M.tuberculosis_H37Rv ADVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQRNVQVL
MLBr_02061|M.leprae_Br4923 ANVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQRNIQVL
MUL_3025|M.ulcerans_Agy99 ADVDIKLIARTTHHLFQPLLYQVATGIISEGDIAPPTRVVLRRQRNAQVL
MMAR_2728|M.marinum_M ADVDIKLIARTTHHLFQPLLYQVATGIISEGDIAPPTRVVLRRQRNAQVL
MAV_2867|M.avium_104 ANVDIKLIARTTHHLFQPLLYQVATGIVSEGDIAPPTRVVLRRQRNVQVL
MAB_2429c|M.abscessus_ATCC_199 ANADVTLIARTTHHLFQPLLYQVATGIISEGEIAPPTRQILKDQDNAQVV
*:.*:.:********************:***:***.** :*: * * **:
Mflv_3348|M.gilvum_PYR-GCK LGDVTRIDLKTNTVRSELLGHTYITPFDSLIVAAGAGQSYFGNDHFAEWA
Mvan_3078|M.vanbaalenii_PYR-1 LGDVTGIDLQTKTVRSELLGHTYVTPFDSLIVAAGAGQSYFGNDHFAEWA
MSMEG_3621|M.smegmatis_MC2_155 LGDVTHIDLENKTVDSVLLGHTYSTPYDSLIIAAGAGQSYFGNDHFAEFA
TH_1173|M.thermoresistible__bu LGDVTRIDLTNRRVESVLLGHTYSTPYDSLIVAAGAGQSYFGNDHFAEFA
Mb1885c|M.bovis_AF2122/97 LGNVTHIDLAGQCVVSELLGHTYQTPYDSLIVAAGAGQSYFGNDHFAEFA
Rv1854c|M.tuberculosis_H37Rv LGNVTHIDLAGQCVVSELLGHTYQTPYDSLIVAAGAGQSYFGNDHFAEFA
MLBr_02061|M.leprae_Br4923 LGNVTHIDLANQCVVSELLGHTYETLYDSLIVAAGAGQSYFGNDQFAEFA
MUL_3025|M.ulcerans_Agy99 LGNVTHIDLAKQSVVSDLLGHTYETPYDTLIVAAGAGQSYFGNDHFAEFA
MMAR_2728|M.marinum_M LGNVTHIDLAKQSVVSDLLGHTYETPYDTLIVAAGAGQSYFGNDHFAEFA
MAV_2867|M.avium_104 LGDVTHIDLAGKFVVSDLLGHTYETPYDTLIVAAGAGQSYFGNDHFAEFA
MAB_2429c|M.abscessus_ATCC_199 LGDVTSIDLEKQVVHSSLLGHDYSTPYDSLIVAAGAGQSYFGNDHFAEWA
**:** *** . * * **** * * :*:**:************:***:*
Mflv_3348|M.gilvum_PYR-GCK PGMKSIDDALELRGRILGSFEQAERSSDPARREKLLTFVVVGAGPTGVEM
Mvan_3078|M.vanbaalenii_PYR-1 PGMKSIDDALELRGRILGAFEQAERSSDPARREKLLTFVVVGAGPTGVEM
MSMEG_3621|M.smegmatis_MC2_155 PGMKSIDDALELRGRILGAFEQAERSSDPVRRAKLLTFTVVGAGPTGVEM
TH_1173|M.thermoresistible__bu PGMKTIDDALELRARILGAFEQAERSSDAKRREKLLTFTVVGAGPTGVEM
Mb1885c|M.bovis_AF2122/97 PGMKSIDDALELRGRILSAFEQAERSSDPERRAKLLTFTVVGAGPTGVEM
Rv1854c|M.tuberculosis_H37Rv PGMKSIDDALELRGRILSAFEQAERSSDPERRAKLLTFTVVGAGPTGVEM
MLBr_02061|M.leprae_Br4923 PGMKSIDDALELRGRILSAFEQAERSNDPERREKLLTFTVVGAGPTGVEM
MUL_3025|M.ulcerans_Agy99 PGMKSIDDALELRGRILSAFEQAERSRDAERRAKLLTFTVVGAGPTGVEM
MMAR_2728|M.marinum_M PGMKSIDDALELRGRILSAFEQAERSRDAERRAKLLTFTVVGAGPTGVEM
MAV_2867|M.avium_104 PGMKSIDDALEVRGRILSAFEQAERSRDPERRAKLLTFTVIGAGPTGVEM
MAB_2429c|M.abscessus_ATCC_199 PGMKSIDDALELRGRILGAFEQAERSSDPVRRRKLMTFTVVGAGPTGVEM
****:******:*.***.:******* *. ** **:**.*:*********
Mflv_3348|M.gilvum_PYR-GCK AGQIAELADHTLKGAFRHIDS-TKARVILLDAAPAVLPPMGEKLGKQAQK
Mvan_3078|M.vanbaalenii_PYR-1 AGQIAELADHTLKGAFRHIDS-TKARVILLDAAPAVLPPMGEKLGKKAQA
MSMEG_3621|M.smegmatis_MC2_155 AGQIAELADQTLRGSFRHIDP-TEARVILLDAAPAVLPPMGEKLGKKARA
TH_1173|M.thermoresistible__bu AGQIAELAKYTLKGTFRSIDPATEARVILLDAAPAVLPTMGEKLGKKAQA
Mb1885c|M.bovis_AF2122/97 AGQIAELAEHTLKGAFRHIDS-TKARVILLDAAPAVLPPMGAKLGQRAAA
Rv1854c|M.tuberculosis_H37Rv AGQIAELAEHTLKGAFRHIDS-TKARVILLDAAPAVLPPMGAKLGQRAAA
MLBr_02061|M.leprae_Br4923 AGQIAELADHTLKGAFRRIDS-TKARVVLLDAAPAVLPPMGEKLGQRAAA
MUL_3025|M.ulcerans_Agy99 AGQIAELAEHTLKGAFRHIDS-TRARVILLDAAPAVLPPFGDKLGERAAA
MMAR_2728|M.marinum_M AGQIAELAEHTLKGAFRHIDS-TRARVILLDAAPAVLPPFGDKLGERAAA
MAV_2867|M.avium_104 AGQIAELATYTLKGSFRHIDP-TKARVILLDAAPAVLPPFGDKLGKRAAD
MAB_2429c|M.abscessus_ATCC_199 AGQIAELANETLKGTFRHIDP-TDARVILLDAAPAVLPPFGPKLGDKARK
******** **:*:** **. * ***:**********.:* ***.:*
Mflv_3348|M.gilvum_PYR-GCK RLEKMGVEIQLNAMVTDVDRNGITVKDPDGTMRRIEAACKVWSAGVSASP
Mvan_3078|M.vanbaalenii_PYR-1 RLEKLGVEIQLNAMVTDVDRNGITVKDPDGTIRRIEAACKVWSAGVSASP
MSMEG_3621|M.smegmatis_MC2_155 RLEKMGVEVQLGAMVTDVDRNGITVKDSDGTIRRIESACKVWSAGVSASP
TH_1173|M.thermoresistible__bu RLEKMGVEIQLGAMVTDVDRNGITVKDSDGTVRRIESACKVWSAGVSASP
Mb1885c|M.bovis_AF2122/97 RLQKLGVEIQLGAMVTDVDRNGITVKDSDGTVRRIESACKVWSAGVSASW
Rv1854c|M.tuberculosis_H37Rv RLQKLGVEIQLGAMVTDVDRNGITVKDSDGTVRRIESACKVWSAGVSASR
MLBr_02061|M.leprae_Br4923 RLQKMGVEIQLSAMVTDVDRNGITVQDSDGTVRRIESACKVWSAGVSASR
MUL_3025|M.ulcerans_Agy99 RLQKLGVEIQLGAMVTDVDRNGITVKDSDGTIRRIESACKVWSAGVQASR
MMAR_2728|M.marinum_M RLQKLGVEIQLGAMVTDVDRNGITVKDSDGTIRRIESACKVWSAGVQASR
MAV_2867|M.avium_104 RLEKMGVEIQLGAMVTDVDRNGITVKDSDGTVRRIESACKVWSAGVSASP
MAB_2429c|M.abscessus_ATCC_199 RLEKLGVEIQLSAMVTDVDRNGLTVKHADGSIERIESWCKVWSAGVSASP
**:*:***:**.**********:**:..**::.***: ********.**
Mflv_3348|M.gilvum_PYR-GCK LGRDLADQSGVELDRAGRVKVLPDLSIPGHPNVFVVGDMAAVEDVPGMAQ
Mvan_3078|M.vanbaalenii_PYR-1 LGRDLAEGSGVELDRAGRVKVLPDLSLPGHPNVFVVGDMAAVEGVPGMAQ
MSMEG_3621|M.smegmatis_MC2_155 LGKDLAEQSGVELDRAGRVKVQPDLTLPGHPNVFVVGDMAAVEGVPGVAQ
TH_1173|M.thermoresistible__bu LGKDLAEQSGAELDRAGRVKVLPDLSLPGHPNVFVVGDMAAVEGVPGMAQ
Mb1885c|M.bovis_AF2122/97 LGRDLAEQSRVELDRAGRVQVLPDLSIPGYPNVFVVGDMAAVEGVPGVAQ
Rv1854c|M.tuberculosis_H37Rv LGRDLAEQSRVELDRAGRVQVLPDLSIPGYPNVFVVGDMAAVEGVPGVAQ
MLBr_02061|M.leprae_Br4923 LGRDLAEQSLVELDWAGRVKVLPDLSIPGYRNVFVVGDMAAIDGVPGVAQ
MUL_3025|M.ulcerans_Agy99 LGRDLAEQSEVELDRAGRVQVLPDLSVPGHPNVFVIGDMAAVEGVPGVAQ
MMAR_2728|M.marinum_M LGRDLAEQSEVELDRAGRVQVLPDLSVPGHPNVFVIGDMAAVEGVPGVAQ
MAV_2867|M.avium_104 LGRDLAEQSTVELDRAGRVKVLPDLSIPGHPNVFVIGDLAAVEGVPGVAQ
MAB_2429c|M.abscessus_ATCC_199 LGKNLAEQSGVELDRAGRVKVGPDLSIPGHPNVFVVGDMMAVDGVPGVAQ
**::**: * .*** ****:* ***::**: ****:**: *::.***:**
Mflv_3348|M.gilvum_PYR-GCK GAIQGGRYAAKAIAAGLKGAEVSEREPFKYFDKGSMATVSRFSAVAKIGP
Mvan_3078|M.vanbaalenii_PYR-1 GAIQGGKYAAKAIAAGLKGADPTEREPFKYFDKGSMATVSRFSAVAKIGP
MSMEG_3621|M.smegmatis_MC2_155 GAIQGGRYAAKIIKREVSGTSPKIRTPFEYFDKGSMATVSRFSAVAKVGP
TH_1173|M.thermoresistible__bu GAIQGARHAAKLIKNELKGADPADREPFQYFDKGSMATVSRFSAVAKIGP
Mb1885c|M.bovis_AF2122/97 GAIQGAKYVASTIKAELAGANPAEREPFQYFDKGSMATVSRFSAVAKIGP
Rv1854c|M.tuberculosis_H37Rv GAIQGAKYVASTIKAELAGANPAEREPFQYFDKGSMATVSRFSAVAKIGP
MLBr_02061|M.leprae_Br4923 GAIQGAKYVANNIKAELGEANPAEREPFQYFDKGSMATVSRFSAVAKIGP
MUL_3025|M.ulcerans_Agy99 GAIQGAKYVATTIKSELAGANAAEREPFQYFDKGSMATVSRFSAVAKIGP
MMAR_2728|M.marinum_M GAIQGAKYVATTIKSELAGANAAEREPFQYFDKGSMATVSRFSAVAKIGP
MAV_2867|M.avium_104 GAIQGAKYVANTIKAELGGADPAEREPFQYFDKGSMATVSRFSAVAKIGP
MAB_2429c|M.abscessus_ATCC_199 GAIQGGRYAAKQIKAELKGRSPAERAPFKFFDKGSMATVSRYSAVVKIGK
*****.::.*. * : . * **::***********:***.*:*
Mflv_3348|M.gilvum_PYR-GCK LEFGGFIAWLSWLVLHLVYLVGFKTKIVTLLSWTVTFLSTKRGQLTITEQ
Mvan_3078|M.vanbaalenii_PYR-1 LEFEGFVAWLAWLVLHLVYLVGFKTKIVTLLSWTVTFLSTKRGQLTITEQ
MSMEG_3621|M.smegmatis_MC2_155 VEFAGFFAWLCWLVLHLVYLVGFKTKIVTLLSWGVTFLSTKRGQLTITEQ
TH_1173|M.thermoresistible__bu LEFGGFIAWLAWLFLHLVYIVGFKARLTTALSWIVTFLGSHRGQLTITEQ
Mb1885c|M.bovis_AF2122/97 VEFSGFIAWLIWLVLHLAYLIGFKTKITTLLSWTVTFLSTRRGQLTITDQ
Rv1854c|M.tuberculosis_H37Rv VEFSGFIAWLIWLVLHLAYLIGFKTKITTLLSWTVTFLSTRRGQLTITDQ
MLBr_02061|M.leprae_Br4923 LEFSGFIAWLMWLVLHLMYLIGFKTKITTLLSWTVTFLSTRRGQLTITEQ
MUL_3025|M.ulcerans_Agy99 VEISGFIAWLAWLVLHLVYLVGFKTKVSTLLSWTVTFLSTRRGQLTITEQ
MMAR_2728|M.marinum_M VEISGFIAWLAWLVLHLVYLVGFKTKVSTLLSWTVTFLSTRRGQLTITEQ
MAV_2867|M.avium_104 LEFSGLFAWFAWLVLHLVYLVGFKTKVSTLLSWTVTFLSTRRGQLTITEQ
MAB_2429c|M.abscessus_ATCC_199 IEFSGFIAWLSWLALHLVYLVGFKNRLVTLILWSIAFVTRSRGQMAITEQ
:*: *:.**: ** *** *::*** :: * : * ::*: ***::**:*
Mflv_3348|M.gilvum_PYR-GCK QAYARIRLEELEEIAASVQDTEKAAS---------
Mvan_3078|M.vanbaalenii_PYR-1 QAYARTRIEELEEIAASVQETERAAS---------
MSMEG_3621|M.smegmatis_MC2_155 QAYARTRIEELEEIAAAVQDTEKAAS---------
TH_1173|M.thermoresistible__bu QAFARTRIEELQEIAASVQDTQKAAS---------
Mb1885c|M.bovis_AF2122/97 QAFARTRLEQLAELAAEAQGSAASAKVAS------
Rv1854c|M.tuberculosis_H37Rv QAFARTRLEQLAELAAEAQGSAASAKVAS------
MLBr_02061|M.leprae_Br4923 QAFARTRLEQLAELTAESQSTAAANRATMRAS---
MUL_3025|M.ulcerans_Agy99 QAFARTRLEQLAELASEAQSTEARQAS--------
MMAR_2728|M.marinum_M QAFARTRLEQLAELASEAQSTEARQAS--------
MAV_2867|M.avium_104 QAFARTRLEQLAVLAAETKRPAARRAS--------
MAB_2429c|M.abscessus_ATCC_199 QAWARTRLDQLEELVAEKNGDEEPTTQPDKEKAAS
**:** *:::* :.: :