For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSRHRVVIIGSGFGGLTAAKALKRVPKGTQVDITLISKTTTHLFQPLLYQVATGILSEGDIAPTTRLILR KQKNVRVLWGDVSAIDLTARTVTSHLMGMDTVTPYDSLIVAAGAQQSYFGHDEYAAFAPGMKSVDDALEL RGRILGAFEAAEVATDPAERRRRLTFVVVGAGPTGVELAGEIVQLAERTLAGAFRTITPSECRVILLDAA PAVLPPMGPKLGLKAQRRLQKMDVEVQLNAMVTAVDYMGITVKEKDGAERRIECACKVWAAGVQASALGA MIAEQSDGTETDRAGRVIVEPDLTVKGHPYVFVVGDLMSVPGVPGMAQGAIQGAKYAAGIIKQAVKGQDN PAERKPFRYVNKGSMALISRYSAVAQVGRVEFAGFIAWLAWLLLHLYYLIGHRNRLAAMFAWGISFLGRT RGQMAITSQMIYARPVVEWLERRAQDGTLAAAEHVDSAEKQAG
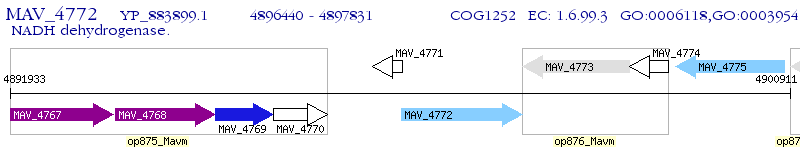
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4772 | - | - | 100% (463) | DoxD family protein/pyridine nucleotide-disulfide oxidoreductase |
| M. avium 104 | MAV_2867 | - | e-154 | 62.44% (450) | DoxD family protein/pyridine nucleotide-disulfide |
| M. avium 104 | MAV_4103 | - | 1e-38 | 30.72% (459) | dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0398c | ndhA | 0.0 | 82.61% (460) | membrane NADH dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3348 | - | 1e-160 | 64.24% (453) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. tuberculosis H37Rv | Rv0392c | ndhA | 0.0 | 82.83% (460) | membrane NADH dehydrogenase |
| M. leprae Br4923 | MLBr_02061 | - | 1e-154 | 62.97% (451) | putative dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2429c | - | 1e-160 | 65.15% (439) | NADH dehydrogenase (NDH) |
| M. marinum M | MMAR_0689 | ndh_1 | 0.0 | 82.83% (460) | NADH dehydrogenase Ndh_1 |
| M. smegmatis MC2 155 | MSMEG_3621 | ndh | 1e-163 | 66.23% (453) | NADH dehydrogenase |
| M. thermoresistible (build 8) | TH_1173 | ndh | 1e-159 | 64.76% (454) | PROBABLE NADH DEHYDROGENASE NDH |
| M. ulcerans Agy99 | MUL_3025 | ndh | 1e-155 | 63.96% (444) | NADH dehydrogenase Ndh |
| M. vanbaalenii PYR-1 | Mvan_3078 | - | 1e-162 | 66.52% (442) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3348|M.gilvum_PYR-GCK ----------------MSHPGATPSD--RHKVVIIGSGFGGLTAAKALKK
Mvan_3078|M.vanbaalenii_PYR-1 ----------------MSHPGATPSD--RHKVVIIGSGFGGLTAAKTLKR
MSMEG_3621|M.smegmatis_MC2_155 ----------------MSHPGATASD--RHKVVIIGSGFGGLTAAKTLKR
TH_1173|M.thermoresistible__bu -----------------------MSD--RHKVVIIGSGFGGLNAAKALRR
MLBr_02061|M.leprae_Br4923 ---------------MNAQPAATAAQDRRHQVVIIGSGFGGLNAAKKLKR
MUL_3025|M.ulcerans_Agy99 ---------------MSPQPETTAQARPRHRVVIIGSGFGGLNAAKKLKR
MAB_2429c|M.abscessus_ATCC_199 MSTTPDAAGAAAPVAANPQPKIQDALTPKHRVVIIGSGFGGLTAAKTLKR
Mb0398c|M.bovis_AF2122/97 ---------------MTLSSGEPSAVGGRHRVVIIGSGFGGLNAAKALKR
Rv0392c|M.tuberculosis_H37Rv ---------------MTLSSGEPSAVGGRHRVVIIGSGFGGLNAAKALKR
MMAR_0689|M.marinum_M ---------------MTPPSSESSAAGRRHHVVIIGSGFGGLSAARALKR
MAV_4772|M.avium_104 ---------------MS-----------RHRVVIIGSGFGGLTAAKALKR
:*:***********.**: *::
Mflv_3348|M.gilvum_PYR-GCK A----DVDIKMIARTTHHLFQPLLYQVATGIISEGEIAPATRVILRNQKN
Mvan_3078|M.vanbaalenii_PYR-1 A----DVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPATRVILRNQKN
MSMEG_3621|M.smegmatis_MC2_155 A----DVDVKLIARTTHHLFQPLLYQVATGIISEGEIAPATRVILRKQKN
TH_1173|M.thermoresistible__bu A----DVDVKLIARTTHHLFQPLLYQVATGIISEGEIAPATRVVLRRQRN
MLBr_02061|M.leprae_Br4923 A----NVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRKQRN
MUL_3025|M.ulcerans_Agy99 A----DVDIKLIARTTHHLFQPLLYQVATGIISEGDIAPPTRVVLRRQRN
MAB_2429c|M.abscessus_ATCC_199 A----NADVTLIARTTHHLFQPLLYQVATGIISEGEIAPPTRQILKDQDN
Mb0398c|M.bovis_AF2122/97 A----DVDITLISKTTTHLFQPLLYQVATGILSEGDIAPTTRLILRRQKN
Rv0392c|M.tuberculosis_H37Rv A----DVDITLISKTTTHLFQPLLYQVATGILSEGDIAPTTRLILRRQKN
MMAR_0689|M.marinum_M A----DVDITLISKTTTHLFQPLLYQVATGILSEGDIAPTTRLILRRQKN
MAV_4772|M.avium_104 VPKGTQVDITLISKTTTHLFQPLLYQVATGILSEGDIAPTTRLILRKQKN
. :.*:.:*::** **************:***:***.** :*: * *
Mflv_3348|M.gilvum_PYR-GCK AQVLLGDVTRIDLKTNTVRSELLGHTYITPFDSLIVAAGAGQSYFGNDHF
Mvan_3078|M.vanbaalenii_PYR-1 AQVLLGDVTGIDLQTKTVRSELLGHTYVTPFDSLIVAAGAGQSYFGNDHF
MSMEG_3621|M.smegmatis_MC2_155 AQVLLGDVTHIDLENKTVDSVLLGHTYSTPYDSLIIAAGAGQSYFGNDHF
TH_1173|M.thermoresistible__bu VQVLLGDVTRIDLTNRRVESVLLGHTYSTPYDSLIVAAGAGQSYFGNDHF
MLBr_02061|M.leprae_Br4923 IQVLLGNVTHIDLANQCVVSELLGHTYETLYDSLIVAAGAGQSYFGNDQF
MUL_3025|M.ulcerans_Agy99 AQVLLGNVTHIDLAKQSVVSDLLGHTYETPYDTLIVAAGAGQSYFGNDHF
MAB_2429c|M.abscessus_ATCC_199 AQVVLGDVTSIDLEKQVVHSSLLGHDYSTPYDSLIVAAGAGQSYFGNDHF
Mb0398c|M.bovis_AF2122/97 VRVLLGEVNAIDLKAQTVTSKLMDMTTVTPYDSLIVAAGAQQSYFGNDEF
Rv0392c|M.tuberculosis_H37Rv VRVLLGEVNAIDLKAQTVTSKLMDMTTVTPYDSLIVAAGAQQSYFGNDEF
MMAR_0689|M.marinum_M VRVLLGEVMGIDLNAQTVTSKLMDMTTVSPYDSLIVAAGAQQSYFGNDDF
MAV_4772|M.avium_104 VRVLWGDVSAIDLTARTVTSHLMGMDTVTPYDSLIVAAGAQQSYFGHDEY
:*: *:* *** . * * *:. : :*:**:**** *****:*.:
Mflv_3348|M.gilvum_PYR-GCK AEWAPGMKSIDDALELRGRILGSFEQAERSSDPARREKLLTFVVVGAGPT
Mvan_3078|M.vanbaalenii_PYR-1 AEWAPGMKSIDDALELRGRILGAFEQAERSSDPARREKLLTFVVVGAGPT
MSMEG_3621|M.smegmatis_MC2_155 AEFAPGMKSIDDALELRGRILGAFEQAERSSDPVRRAKLLTFTVVGAGPT
TH_1173|M.thermoresistible__bu AEFAPGMKTIDDALELRARILGAFEQAERSSDAKRREKLLTFTVVGAGPT
MLBr_02061|M.leprae_Br4923 AEFAPGMKSIDDALELRGRILSAFEQAERSNDPERREKLLTFTVVGAGPT
MUL_3025|M.ulcerans_Agy99 AEFAPGMKSIDDALELRGRILSAFEQAERSRDAERRAKLLTFTVVGAGPT
MAB_2429c|M.abscessus_ATCC_199 AEWAPGMKSIDDALELRGRILGAFEQAERSSDPVRRRKLMTFTVVGAGPT
Mb0398c|M.bovis_AF2122/97 ATFAPGMKTIDDALELRGRILGAFEAAEVSTDHAERERRLTFVVVGAGPT
Rv0392c|M.tuberculosis_H37Rv ATFAPGMKTIDDALELRGRILGAFEAAEVSTDHAERERRLTFVVVGAGPT
MMAR_0689|M.marinum_M AIFAPGMKTIDDALELRGRILGAFEAAEVATDHDERERRLTFVVVGAGPT
MAV_4772|M.avium_104 AAFAPGMKSVDDALELRGRILGAFEAAEVATDPAERRRRLTFVVVGAGPT
* :*****::*******.***.:** ** : * .* : :**.*******
Mflv_3348|M.gilvum_PYR-GCK GVEMAGQIAELADHTLKGAFRHID-STKARVILLDAAPAVLPPMGEKLGK
Mvan_3078|M.vanbaalenii_PYR-1 GVEMAGQIAELADHTLKGAFRHID-STKARVILLDAAPAVLPPMGEKLGK
MSMEG_3621|M.smegmatis_MC2_155 GVEMAGQIAELADQTLRGSFRHID-PTEARVILLDAAPAVLPPMGEKLGK
TH_1173|M.thermoresistible__bu GVEMAGQIAELAKYTLKGTFRSIDPATEARVILLDAAPAVLPTMGEKLGK
MLBr_02061|M.leprae_Br4923 GVEMAGQIAELADHTLKGAFRRID-STKARVVLLDAAPAVLPPMGEKLGQ
MUL_3025|M.ulcerans_Agy99 GVEMAGQIAELAEHTLKGAFRHID-STRARVILLDAAPAVLPPFGDKLGE
MAB_2429c|M.abscessus_ATCC_199 GVEMAGQIAELANETLKGTFRHID-PTDARVILLDAAPAVLPPFGPKLGD
Mb0398c|M.bovis_AF2122/97 GVEVAGQIVELAERTLAGAFRTIT-PSECRVILLDAAPAVLPPMGPKLGL
Rv0392c|M.tuberculosis_H37Rv GVEVAGQIVELAERTLAGAFRTIT-PSECRVILLDAAPAVLPPMGPKLGL
MMAR_0689|M.marinum_M GVELAGEIVQLAERTLAGAFRTIT-PSECRVILLDAAPAVLPPMGPKLGL
MAV_4772|M.avium_104 GVELAGEIVQLAERTLAGAFRTIT-PSECRVILLDAAPAVLPPMGPKLGL
***:**:*.:**. ** *:** * .: .**:**********.:* ***
Mflv_3348|M.gilvum_PYR-GCK QAQKRLEKMGVEIQLNAMVTDVDRNGITVKDPDGTMRRIEAACKVWSAGV
Mvan_3078|M.vanbaalenii_PYR-1 KAQARLEKLGVEIQLNAMVTDVDRNGITVKDPDGTIRRIEAACKVWSAGV
MSMEG_3621|M.smegmatis_MC2_155 KARARLEKMGVEVQLGAMVTDVDRNGITVKDSDGTIRRIESACKVWSAGV
TH_1173|M.thermoresistible__bu KAQARLEKMGVEIQLGAMVTDVDRNGITVKDSDGTVRRIESACKVWSAGV
MLBr_02061|M.leprae_Br4923 RAAARLQKMGVEIQLSAMVTDVDRNGITVQDSDGTVRRIESACKVWSAGV
MUL_3025|M.ulcerans_Agy99 RAAARLQKLGVEIQLGAMVTDVDRNGITVKDSDGTIRRIESACKVWSAGV
MAB_2429c|M.abscessus_ATCC_199 KARKRLEKLGVEIQLSAMVTDVDRNGLTVKHADGSIERIESWCKVWSAGV
Mb0398c|M.bovis_AF2122/97 KAQRRLEKMDAEVQLNAMVTAVDYKGITIKEKDGGERRIECACKVWAAGV
Rv0392c|M.tuberculosis_H37Rv KAQRRLEKMDVEVQLNAMVTAVDYKGITIKEKDGGERRIECACKVWAAGV
MMAR_0689|M.marinum_M KAQKKLEKMDVEIQLNAMVTAVDYKGITVKDKDGTERRIDCACKVWAAGV
MAV_4772|M.avium_104 KAQRRLQKMDVEVQLNAMVTAVDYMGITVKEKDGAERRIECACKVWAAGV
:* :*:*:..*:**.**** ** *:*::. ** .**:. ****:***
Mflv_3348|M.gilvum_PYR-GCK SASPLGRDLADQS-GVELDRAGRVKVLPDLSIPGHPNVFVVGDMAAVEDV
Mvan_3078|M.vanbaalenii_PYR-1 SASPLGRDLAEGS-GVELDRAGRVKVLPDLSLPGHPNVFVVGDMAAVEGV
MSMEG_3621|M.smegmatis_MC2_155 SASPLGKDLAEQS-GVELDRAGRVKVQPDLTLPGHPNVFVVGDMAAVEGV
TH_1173|M.thermoresistible__bu SASPLGKDLAEQS-GAELDRAGRVKVLPDLSLPGHPNVFVVGDMAAVEGV
MLBr_02061|M.leprae_Br4923 SASRLGRDLAEQS-LVELDWAGRVKVLPDLSIPGYRNVFVVGDMAAIDGV
MUL_3025|M.ulcerans_Agy99 QASRLGRDLAEQS-EVELDRAGRVQVLPDLSVPGHPNVFVIGDMAAVEGV
MAB_2429c|M.abscessus_ATCC_199 SASPLGKNLAEQS-GVELDRAGRVKVGPDLSIPGHPNVFVVGDMMAVDGV
Mb0398c|M.bovis_AF2122/97 AASPLGKMIAEGSDGTEIDRAGRVIVEPDLTVKGHPNVFVVGDLMFVPGV
Rv0392c|M.tuberculosis_H37Rv AASPLGKMIAEGSDGTEIDRAGRVIVEPDLTVKGHPNVFVVGDLMFVPGV
MMAR_0689|M.marinum_M QASPLGKMIAEQSDGTETDRAGRVIVEPDLTVKGHPHVFVIGDLMSVPGV
MAV_4772|M.avium_104 QASALGAMIAEQSDGTETDRAGRVIVEPDLTVKGHPYVFVVGDLMSVPGV
** ** :*: * .* * **** * ***:: *: ***:**: : .*
Mflv_3348|M.gilvum_PYR-GCK PGMAQGAIQGGRYAAKAIAAGLKGAE-VSEREPFKYFDKGSMATVSRFSA
Mvan_3078|M.vanbaalenii_PYR-1 PGMAQGAIQGGKYAAKAIAAGLKGAD-PTEREPFKYFDKGSMATVSRFSA
MSMEG_3621|M.smegmatis_MC2_155 PGVAQGAIQGGRYAAKIIKREVSGTS-PKIRTPFEYFDKGSMATVSRFSA
TH_1173|M.thermoresistible__bu PGMAQGAIQGARHAAKLIKNELKGAD-PADREPFQYFDKGSMATVSRFSA
MLBr_02061|M.leprae_Br4923 PGVAQGAIQGAKYVANNIKAELGEAN-PAEREPFQYFDKGSMATVSRFSA
MUL_3025|M.ulcerans_Agy99 PGVAQGAIQGAKYVATTIKSELAGAN-AAEREPFQYFDKGSMATVSRFSA
MAB_2429c|M.abscessus_ATCC_199 PGVAQGAIQGGRYAAKQIKAELKGRS-PAERAPFKFFDKGSMATVSRYSA
Mb0398c|M.bovis_AF2122/97 PGVAQGAIQGARYATTVIKHMVKGNDDPANRKPFHYFNKGSMATISRHSA
Rv0392c|M.tuberculosis_H37Rv PGVAQGAIQGARYATTVIKHMVKGNDDPANRKPFHYFNKGSMATISRHSA
MMAR_0689|M.marinum_M PGMAQGAIQGAHYATKTIKQAVKGHDDPANRKPFQYFNKGSMALISHYSA
MAV_4772|M.avium_104 PGMAQGAIQGAKYAAGIIKQAVKGQDNPAERKPFRYVNKGSMALISRYSA
**:*******.::.: * : . * **.:.:***** :*:.**
Mflv_3348|M.gilvum_PYR-GCK VAKIGPLEFGGFIAWLSWLVLHLVYLVGFKTKIVTLLSWTVTFLSTKRGQ
Mvan_3078|M.vanbaalenii_PYR-1 VAKIGPLEFEGFVAWLAWLVLHLVYLVGFKTKIVTLLSWTVTFLSTKRGQ
MSMEG_3621|M.smegmatis_MC2_155 VAKVGPVEFAGFFAWLCWLVLHLVYLVGFKTKIVTLLSWGVTFLSTKRGQ
TH_1173|M.thermoresistible__bu VAKIGPLEFGGFIAWLAWLFLHLVYIVGFKARLTTALSWIVTFLGSHRGQ
MLBr_02061|M.leprae_Br4923 VAKIGPLEFSGFIAWLMWLVLHLMYLIGFKTKITTLLSWTVTFLSTRRGQ
MUL_3025|M.ulcerans_Agy99 VAKIGPVEISGFIAWLAWLVLHLVYLVGFKTKVSTLLSWTVTFLSTRRGQ
MAB_2429c|M.abscessus_ATCC_199 VVKIGKIEFSGFIAWLSWLALHLVYLVGFKNRLVTLILWSIAFVTRSRGQ
Mb0398c|M.bovis_AF2122/97 VAQVGKLEFAGYFAWLAWLVLHLVYLVGYRNRIAALFAWGISFMGRARGQ
Rv0392c|M.tuberculosis_H37Rv VAQVGKLEFAGYFAWLAWLVLHLVYLVGYRNRIAALFAWGISFMGRARGQ
MMAR_0689|M.marinum_M VAKVGKVEFAGFIAWLAWLFLHLLYLVGHRNRVAAAFSWGLAFLGRTRGQ
MAV_4772|M.avium_104 VAQVGRVEFAGFIAWLAWLLLHLYYLIGHRNRLAAMFAWGISFLGRTRGQ
*.::* :*: *:.*** ** *** *::*.: :: : : * ::*: ***
Mflv_3348|M.gilvum_PYR-GCK LTITEQQAYARIRLEELEEIAASVQDTEKA---------AS
Mvan_3078|M.vanbaalenii_PYR-1 LTITEQQAYARTRIEELEEIAASVQETERA---------AS
MSMEG_3621|M.smegmatis_MC2_155 LTITEQQAYARTRIEELEEIAAAVQDTEKA---------AS
TH_1173|M.thermoresistible__bu LTITEQQAFARTRIEELQEIAASVQDTQKA---------AS
MLBr_02061|M.leprae_Br4923 LTITEQQAFARTRLEQLAELTAESQSTAAANRAT---MRAS
MUL_3025|M.ulcerans_Agy99 LTITEQQAFARTRLEQLAELASEAQSTEAR--------QAS
MAB_2429c|M.abscessus_ATCC_199 MAITEQQAWARTRLDQLEELVAEKNGDEEPTTQPDKEKAAS
Mb0398c|M.bovis_AF2122/97 MAITSQMIYARLVMTLMEQQAQG-ALAAAEQAEHAEQEAAG
Rv0392c|M.tuberculosis_H37Rv MAITSQMIYARLVMTLMEQQAQG-ALAAAEQAEHAEQEAAG
MMAR_0689|M.marinum_M MAITSQMVYARLAMRYLQARAEEDALSAAEHAEQAEQRATG
MAV_4772|M.avium_104 MAITSQMIYARPVVEWLERRAQDGTLAAAEHVDSAEKQAG-
::**.* :** : : .