For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGRLAGRAAIVTGASRGLGRAIALALASEGAAVAVAGRTEQVWDDRLPGTIGSTVADIEAAGGRAVPVRA DLTDRDDVARLVDSAREALGPITILVNNAAFTAPGRPPAPGGEARAKPAAGGEARAKPASGGAKPASGGA KPASGGAKPASGGAKPGWPGFVSTPLHAYRRHFDIAVFAAYELMQRVCPDMIGAGGGAIINITSVASRLP GDGPYADRSGGVLPGYGGSKAALEHLTQCVAYDLADHRIAVNALSPSKPILTPGLSYYARDFDDTASADE FARAAVELALVDPGRVTGRTIGHLQVLDGSFRPSGWTRLWRLGLGSSGPHHRGGGGVAPSPTAAHGFSDP EARQVQVRQVRPVRRADPPAPERRVRPARREVLPPPARREVLPPLARRVAPPRPERPALREVRAAREDRA VQVARVAAGCSAGSSPRGRLRCPASRGTRLPTRPPRRVLR*
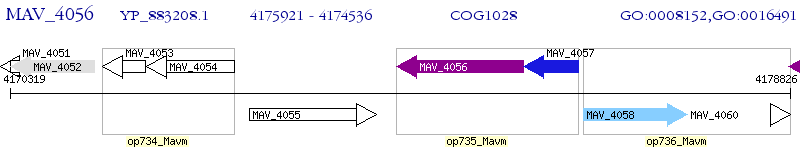
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4056 | - | - | 100% (461) | oxidoreductase, short chain dehydrogenase/reductase family protein |
| M. avium 104 | MAV_1046 | - | 1e-20 | 30.77% (312) | short chain dehydrogenase |
| M. avium 104 | MAV_3027 | - | 5e-19 | 50.48% (105) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0950c | - | 2e-18 | 29.81% (312) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2396 | - | 1e-21 | 31.33% (300) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0927c | - | 2e-18 | 29.81% (312) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 2e-08 | 31.68% (101) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4632 | - | 9e-19 | 29.59% (267) | short chain dehydrogenase |
| M. marinum M | MMAR_1395 | - | 1e-127 | 75.32% (316) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_5584 | - | 3e-22 | 32.38% (315) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3898 | - | 1e-21 | 31.38% (325) | PUTATIVE short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_2481 | - | 1e-110 | 73.31% (281) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_4921 | - | 7e-21 | 32.27% (313) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1395|M.marinum_M -------------MTGALAGRAAIVTGASRGLGRAIALALAAEGAAVVVA
MUL_2481|M.ulcerans_Agy99 ------------------------------------------------MA
MAV_4056|M.avium_104 -------------MTGRLAGRAAIVTGASRGLGRAIALALASEGAAVAVA
Mflv_2396|M.gilvum_PYR-GCK ---------------MTLDGKVAFVAGASRGIGATVAAALARAGASVAVA
TH_3898|M.thermoresistible__bu ----------VILDRFRLDDRVAVVTGAGRGLGAAMAVAFAEAGADVLIS
Mvan_4921|M.vanbaalenii_PYR-1 MTRGRQREANVILDRFRLDDQVAVVTGAGRGLGAAMALAFAEAGADVLIA
MSMEG_5584|M.smegmatis_MC2_155 ---------MLILDRFRLDDKVAVVTGAGRGLGAAIALAFAEAGADVVIA
Mb0950c|M.bovis_AF2122/97 ----------MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLIA
Rv0927c|M.tuberculosis_H37Rv ----------MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLIA
MAB_4632|M.abscessus_ATCC_1997 ----------MILDRFKLDGNVAVVTGAGRGLGAAIAEAFAQAGADVLIA
MLBr_01094|M.leprae_Br4923 --------------MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAIS
::
MMAR_1395|M.marinum_M GRTEQVWDSRLPGTIGDTVAQIEAAGGRAVAVRADLTDRDDVATLVTGAR
MUL_2481|M.ulcerans_Agy99 GRTEQVWDSRLPGTIGDTVAQIEAAGGRAVAVRADLTDRDDVAALVTAAR
MAV_4056|M.avium_104 GRTEQVWDDRLPGTIGSTVADIEAAGGRAVPVRADLTDRDDVARLVDSAR
Mflv_2396|M.gilvum_PYR-GCK ARTEQ--EGRLPGTIGSVAQRITDEGGRALPVVCDVTREESVEAAVAATV
TH_3898|M.thermoresistible__bu ARTQA--------QLEEVAAQIAATGRRAHVVAADLAHPEATAELAGAAV
Mvan_4921|M.vanbaalenii_PYR-1 ARTRS--------QLEEVAEQIEGTGRRAHIVVADLAHAEDTAALAAGAV
MSMEG_5584|M.smegmatis_MC2_155 ARTQS--------QLDEVAGQVAGAGRKAHVVVADLAHPDATAELVQAAI
Mb0950c|M.bovis_AF2122/97 SRTSS--------ELDAVAEQIRAAGRRAHTVAADLAHPEVTAQLAGQAV
Rv0927c|M.tuberculosis_H37Rv SRTSS--------ELDAVAEQIRAAGRRAHTVAADLAHPEVTAQLAGQAV
MAB_4632|M.abscessus_ATCC_1997 SRTES--------QLREVAAKVQAAGRQAEVVVADLSDTEASAALAQKAV
MLBr_01094|M.leprae_Br4923 DVDGE--------GLAQTEGQLKAIGASARTDRLDVTEREAFLTYADVVH
: . : * * *:: : . .
MMAR_1395|M.marinum_M DSLGPITILVNNAAFTAPGRPPVPGAAPRAK--------------SSRTA
MUL_2481|M.ulcerans_Agy99 DSLGPITILVNNAAFTAPGRPPVPGAAPRAK--------------SSRAA
MAV_4056|M.avium_104 EALGPITILVNNAAFTAPGRPPAPGGEARAKPAAGGEARAKPASGGAKPA
Mflv_2396|M.gilvum_PYR-GCK AEFGGIDILVANAGVLWLG-------------------------------
TH_3898|M.thermoresistible__bu EAFGKLDIVVNNVGGTMPG-------------------------------
Mvan_4921|M.vanbaalenii_PYR-1 EAFGRLDIVVNNVGGTMPN-------------------------------
MSMEG_5584|M.smegmatis_MC2_155 EAFGKLDVVVNNVGGTMPA-------------------------------
Mb0950c|M.bovis_AF2122/97 GAFGKLDIVVNNVGGTMPN-------------------------------
Rv0927c|M.tuberculosis_H37Rv GAFGKLDIVVNNVGGTMPN-------------------------------
MAB_4632|M.abscessus_ATCC_1997 DRFGRLDVVVNNVGGTMPK-------------------------------
MLBr_01094|M.leprae_Br4923 ENFGKVNQIYNNAGIAFTG-------------------------------
:* : : *..
MMAR_1395|M.marinum_M VG----------------KPGWPGFVSVPLAAYRRHFETSVFAAYELMQL
MUL_2481|M.ulcerans_Agy99 VG----------------KPGWPGFVSVPLAAYRRHFETSVFAAYELMQL
MAV_4056|M.avium_104 SGGAKPASGGAKPASGGAKPGWPGFVSTPLHAYRRHFDIAVFAAYELMQR
Mflv_2396|M.gilvum_PYR-GCK -----------------------PIESTPLKRWQLCLDVNTTGVFLVTKA
TH_3898|M.thermoresistible__bu -----------------------PLLNTSAKEMRDAFTFNVGTAHALTTA
Mvan_4921|M.vanbaalenii_PYR-1 -----------------------ALLTTSTKDMRDAFAFNVATAHALTTA
MSMEG_5584|M.smegmatis_MC2_155 -----------------------PLLNTSTKDLRDAFTFNVSTAHALTSA
Mb0950c|M.bovis_AF2122/97 -----------------------TLLSTSTKDLADAFAFNVGTAHALTVA
Rv0927c|M.tuberculosis_H37Rv -----------------------TLLSTSTKDLADAFAFNVGTAHALTVA
MAB_4632|M.abscessus_ATCC_1997 -----------------------PFLDTTNDDLAEAFSFNVLSGHALLRA
MLBr_01094|M.leprae_Br4923 -----------------------DVEVSHFKDIERVMDVDYWGVVNGTKA
. :
MMAR_1395|M.marinum_M SCPDMIA-AGAGSIINITSVASRLPGDGPYPDRSGGVLPGYGGSKAALEH
MUL_2481|M.ulcerans_Agy99 SCPDMIA-AGAGSIINITSVASRLPGDGPYPDRSGGVLPGYGGSKAALEH
MAV_4056|M.avium_104 VCPDMIG-AGGGAIINITSVASRLPGDGPYADRSGGVLPGYGGSKAALEH
Mflv_2396|M.gilvum_PYR-GCK VIPHVRA-RGGGSLIAVTTTGVTMTDHGAN---------AYWVSKAAAER
TH_3898|M.thermoresistible__bu AVPLMLKHSGGGSIINITSTMGRLAGRGFA---------AYGTAKAALAH
Mvan_4921|M.vanbaalenii_PYR-1 AVPLMLEHSGGGSIINITSTMGRLAGRGFA---------AYGTAKAALAH
MSMEG_5584|M.smegmatis_MC2_155 AVPLMLEHSGGGSIINITSTMGRLAGRGFV---------AYGTAKAALAH
Mb0950c|M.bovis_AF2122/97 AVPLMLEHSGGGSVINISSTMGRLAARGFA---------AYGTAKAALAH
Rv0927c|M.tuberculosis_H37Rv AVPLMLEHSGGGSVINISSTMGRLAARGFA---------AYGTAKAALAH
MAB_4632|M.abscessus_ATCC_1997 AVPHLLK-SDNASVINITSTMGRLPGRAFL---------GYCTAKGALAH
MLBr_01094|M.leprae_Br4923 FLPYLIS-SGDGHVINISSVFGLFSVPGQA---------AYNSAKFAVRG
* : . . :* :::. :. . .* :* *
MMAR_1395|M.marinum_M LTQCAAFDLADH--HIAVNALAPSKPIPTPGLSYYAR------------E
MUL_2481|M.ulcerans_Agy99 LTQCAAFDLADH--HIAVNALAPSKPIPTPGLSYYAR------------E
MAV_4056|M.avium_104 LTQCVAYDLADH--RIAVNALSPSKPILTPGLSYYAR------------D
Mflv_2396|M.gilvum_PYR-GCK LYLGLAADLKPD--NIAVNCLSPSRVVLTEGWQAGGAGMEI--------P
TH_3898|M.thermoresistible__bu YTRLAALDLAP---RIRVNAIAPG-SILTSALDVVASNDELRTPMEQATP
Mvan_4921|M.vanbaalenii_PYR-1 YTRLSALDLAP---RIRVNAIAPG-SILTSALEIVADNDELRGPMEKATP
MSMEG_5584|M.smegmatis_MC2_155 YTRLAALDLCP---RIRVNAIAPG-SILTSALDVVASNDALREPMEKVTP
Mb0950c|M.bovis_AF2122/97 YTRLAALDLCP---RVRVNAIAPG-SILTSALEVVAANDELRAPMEQATP
Rv0927c|M.tuberculosis_H37Rv YTRLAALDLCP---RVRVNAIAPG-SILTSALEVVAANDELRAPMEQATP
MAB_4632|M.abscessus_ATCC_1997 YTRTAAMDLSP---RIRVNGIAPG-SILTSALEVVAGNDEMRNTLEQNTP
MLBr_01094|M.leprae_Br4923 FTEALREEMALAGRPVNVTTVYPGGIKTAIARNATAAEGLDVSKIASRFD
:: : *. : *. : . . .
MMAR_1395|M.marinum_M FDDTASAEEFARAAVVLASVD--ADLVTGRTIGHHEVLDGSFRAFGRD--
MUL_2481|M.ulcerans_Agy99 FDDTASAEEFARAAVVLASVD--ADLVTGRTIGHHEVLDGSFRAFGRD--
MAV_4056|M.avium_104 FDDTASADEFARAAVELALVD--PGRVTGRTIGHLQVLDGSFRPSGWTRL
Mflv_2396|M.gilvum_PYR-GCK PEMVEPPEAMGRATVMLAQQD--ASGVTG-TIQRSEALAL----------
TH_3898|M.thermoresistible__bu LRRLGEPADIAAAAVYLASPA--GSYLTGKVLEVDGGLTRPNLDMPIPDL
Mvan_4921|M.vanbaalenii_PYR-1 LRRLGDPLDIAAAAVYLASPA--GSYLTGKTLEVDGGITFPNLDLPIPDL
MSMEG_5584|M.smegmatis_MC2_155 LRRLGDPADIAAAAVYLASPA--GAYLTGKTLEVDGGLTMPNLDLPIPDL
Mb0950c|M.bovis_AF2122/97 LRRLGDPVDIAAAAVYLASPA--GSFLTGKTLEVDGGLTFPNLDLPIPDL
Rv0927c|M.tuberculosis_H37Rv LRRLGDPVDIAAAAVYLASPA--GSFLTGKTLEVDGGLTFPNLDLPIPDL
MAB_4632|M.abscessus_ATCC_1997 LHRLGDPADIAAAAVYLASPA--GSFLTGKVLEVDGGLIAPNLDIPLPDL
MLBr_01094|M.leprae_Br4923 TWVAHTSPQHAARIILKAVRKKKARVLVGPDAKVANVVVRFSGGAGYQRL
. . : * :.* :
MMAR_1395|M.marinum_M --------------------------------------------------
MUL_2481|M.ulcerans_Agy99 --------------------------------------------------
MAV_4056|M.avium_104 WRLGLGSSGPHHRGGGGVAPSPTAAHGFSDPEARQVQVRQVRPVRRADPP
Mflv_2396|M.gilvum_PYR-GCK --------------------------------------------------
TH_3898|M.thermoresistible__bu --------------------------------------------------
Mvan_4921|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5584|M.smegmatis_MC2_155 --------------------------------------------------
Mb0950c|M.bovis_AF2122/97 --------------------------------------------------
Rv0927c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4632|M.abscessus_ATCC_1997 A-------------------------------------------------
MLBr_01094|M.leprae_Br4923 FAQVASRLILNQR-------------------------------------
MMAR_1395|M.marinum_M --------------------------------------------------
MUL_2481|M.ulcerans_Agy99 --------------------------------------------------
MAV_4056|M.avium_104 APERRVRPARREVLPPPARREVLPPLARRVAPPRPERPALREVRAAREDR
Mflv_2396|M.gilvum_PYR-GCK --------------------------------------------------
TH_3898|M.thermoresistible__bu --------------------------------------------------
Mvan_4921|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5584|M.smegmatis_MC2_155 --------------------------------------------------
Mb0950c|M.bovis_AF2122/97 --------------------------------------------------
Rv0927c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4632|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
MMAR_1395|M.marinum_M -----------------------------------------
MUL_2481|M.ulcerans_Agy99 -----------------------------------------
MAV_4056|M.avium_104 AVQVARVAAGCSAGSSPRGRLRCPASRGTRLPTRPPRRVLR
Mflv_2396|M.gilvum_PYR-GCK -----------------------------------------
TH_3898|M.thermoresistible__bu -----------------------------------------
Mvan_4921|M.vanbaalenii_PYR-1 -----------------------------------------
MSMEG_5584|M.smegmatis_MC2_155 -----------------------------------------
Mb0950c|M.bovis_AF2122/97 -----------------------------------------
Rv0927c|M.tuberculosis_H37Rv -----------------------------------------
MAB_4632|M.abscessus_ATCC_1997 -----------------------------------------
MLBr_01094|M.leprae_Br4923 -----------------------------------------