For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AASDRHPQRLTPLPADEWDEDTRAALASLIPAERANPVGAGNVLSTLVRHPDLTAAYLPFNAYLLTRSTL SPRIREVALLRVVHRSKCGYLWSHHLPIARRAGLSESDIDDIRRGRCADPTDAAVVRAVDELTGESTLSR QTWDRLCELFTQRQCMDLIFTIGGYLLLALAVNTFGVQEEEETR*
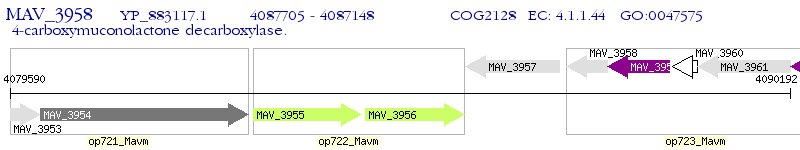
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3958 | pcaC | - | 100% (185) | hypothetical protein MAV_3958 |
| M. avium 104 | MAV_0906 | pcaC | 3e-41 | 50.28% (177) | hypothetical protein MAV_0906 |
| M. avium 104 | MAV_1392 | pcaC | 3e-22 | 33.87% (186) | hypothetical protein MAV_1392 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1558 | - | 5e-11 | 25.68% (148) | hypothetical protein Mb1558 |
| M. gilvum PYR-GCK | Mflv_2473 | - | 2e-52 | 58.14% (172) | carboxymuconolactone decarboxylase |
| M. tuberculosis H37Rv | Rv1531 | - | 5e-11 | 25.68% (148) | hypothetical protein Rv1531 |
| M. leprae Br4923 | MLBr_02465 | - | 8e-06 | 24.78% (113) | hypothetical protein MLBr_02465 |
| M. abscessus ATCC 19977 | MAB_4119 | - | 3e-10 | 29.03% (124) | hypothetical protein MAB_4119 |
| M. marinum M | MMAR_4743 | - | 8e-48 | 52.60% (173) | hypothetical protein MMAR_4743 |
| M. smegmatis MC2 155 | MSMEG_4839 | - | 1e-51 | 55.81% (172) | carboxymuconolactone decarboxylase |
| M. thermoresistible (build 8) | TH_3878 | - | 2e-52 | 59.30% (172) | PUTATIVE Carboxymuconolactone decarboxylase |
| M. ulcerans Agy99 | MUL_4533 | - | 5e-06 | 29.91% (117) | hypothetical protein MUL_4533 |
| M. vanbaalenii PYR-1 | Mvan_4181 | - | 3e-54 | 60.82% (171) | carboxymuconolactone decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1558|M.bovis_AF2122/97 -----MTTSRVPLLPVDEAKAAADEAGVPDYMAELS-------IFQVLLN
Rv1531|M.tuberculosis_H37Rv -----MTTSRVPLLPVDEAKAAADEAGVPDYMAELS-------IFQVLLN
Mflv_2473|M.gilvum_PYR-GCK -----MSTPRLEPLPADRWDDAARDAVSPLLSEERRNPRGAGNVVGTLAR
Mvan_4181|M.vanbaalenii_PYR-1 ----------MSPLPTEEWDDEVLDALAPLLPPSRSNPRDAGNVLATLVR
TH_3878|M.thermoresistible__bu -------VTRLSPLPAEEWTDEVRRSLAPLLAPDRANPRDAGNVLATLVR
MAV_3958|M.avium_104 MAASDRHPQRLTPLPADEWDEDTRAALASLIPAERANPVGAGNVLSTLVR
MMAR_4743|M.marinum_M --------MRLPPLPADQWDEAVQRSLSVILPAQRCNPRDAGNALSTFAN
MSMEG_4839|M.smegmatis_MC2_155 --------MRLTPLPADEWDDEVRLALSVMLPEERLNPEGAGTALSTLAR
MLBr_02465|M.leprae_Br4923 -MSGGTGTGPVGRIPPGSLRQLGPINWVIAKLAASLLRTSEMHLFTILGQ
MUL_4533|M.ulcerans_Agy99 -MTG----DQVARIPSGGIRQLGPVNWALAKLAARSVRAPEMHLFTALGY
MAB_4119|M.abscessus_ATCC_1997 -MSS-------GRIPSGGLRELGPINWVIAKGMARAVNAPEMHLATTLGQ
:* :
Mb1558|M.bovis_AF2122/97 HPRLARTFNDLLATMLWHGTLDSRLRELVIMRIGWLTDCDYEWTQHWRVA
Rv1531|M.tuberculosis_H37Rv HPRLARTFNDLLATMLWHGTLDSRLRELVIMRIGWLTDCDYEWTQHWRVA
Mflv_2473|M.gilvum_PYR-GCK HPGLTRAYLTFNAHLLLASTLTPRIREVALLRAVHVRPCDYLWDHHLPIA
Mvan_4181|M.vanbaalenii_PYR-1 HPALTRAYLHFNAHLLLASTLSPRIREVALLRAVHVRRSDYLWEHHIPIA
TH_3878|M.thermoresistible__bu HPALTRAYLEFNAYLLRDSTLSTRTREVAVLRVVLLRDCEYLWEHHIPLA
MAV_3958|M.avium_104 HPDLTAAYLPFNAYLLTRSTLSPRIREVALLRVVHRSKCGYLWSHHLPIA
MMAR_4743|M.marinum_M HPALAKAFLRFNTHLLFASTLPARVRELAILRVAHRRACAYEWTHHVALA
MSMEG_4839|M.smegmatis_MC2_155 HPRLTKAFLRFSNHLLFRSTLDPRLRELAILRIARRRNCEYEWAHHAFIG
MLBr_02465|M.leprae_Br4923 RQLLFWAWLIYGGRLLRG-KLPRVDTELVILRVAHLRTCEYELQHHRRMA
MUL_4533|M.ulcerans_Agy99 RQYLFWTWAIYSGRLLHG-RLPRIDTELVILRVAHLRSCEYELQHHRRMA
MAB_4119|M.abscessus_ATCC_1997 TGARFWPWLAYSGAILRGTKLSTRDTEVVILRVAHVRECEYELQHHTRIA
.: :* * *:.::* . * :* :.
Mb1558|M.bovis_AF2122/97 SGLGVSADDLLGVRDW---QGYNG----FGPAEQAVLAATDDVVREGAVS
Rv1531|M.tuberculosis_H37Rv SGLGVSADDLLGVRDW---QGYNG----FGPAEQAVLAATDDVVREGAVS
Mflv_2473|M.gilvum_PYR-GCK HGTGLTDAEIASIRSG---DPDD-------AVDRLVVHAVDELHGSNTLS
Mvan_4181|M.vanbaalenii_PYR-1 QRAGITPDEIDAIRQG---RPAD-------EIDRLVVGAVDELDAANTVS
TH_3878|M.thermoresistible__bu QRAGLTPAEIVGIRNG---RMAD-------EPDQLVVRAADELARDHEIS
MAV_3958|M.avium_104 RRAGLSESDIDDIRRG---RCAD-------PTDAAVVRAVDELTGESTLS
MMAR_4743|M.marinum_M EQAGISGDEIAAVRRG---EATD-------DFERAVLTGVDELDENSELS
MSMEG_4839|M.smegmatis_MC2_155 KAEGLTDDDIDGIGRG---VVSD-------PFDQVVIDAVDELDELSTIS
MLBr_02465|M.leprae_Br4923 RKRGLDTKIQAMIFAWPDVPTGAG----LSVRQQALLAATDEFVKDRKIT
MUL_4533|M.ulcerans_Agy99 RAAGLDAHAQATIFAWPDAPQGDGPRRVLSSRQQALLTATDELIKNRSVT
MAB_4119|M.abscessus_ATCC_1997 KSAGIDPAYQERIFTG---AGAEG----LSDKERALITGVDEILTTRTLS
*: : : :: ..*:. ::
Mb1558|M.bovis_AF2122/97 AQSWSACERELHCDKVVLIELVTVISAWRMVASILHSLEVPLEDGVSSWP
Rv1531|M.tuberculosis_H37Rv AQSWSACERELHCDKVVLIELVTVISAWRMVASILHSLEVPLEDGVSSWP
Mflv_2473|M.gilvum_PYR-GCK DDTWTALS--DHFDERQVLDLIFTVGCYQMLGAAVNALGIQPETH-----
Mvan_4181|M.vanbaalenii_PYR-1 DATWSALR--DHFDERQVLDLIFTVGCYQLLGAAVNTLGVQPEPH-----
TH_3878|M.thermoresistible__bu EGTWRALG--RHFDDRGRMDLVFTVGCYALLAVAVNTFGVEPEHA-----
MAV_3958|M.avium_104 RQTWDRLC--ELFTQRQCMDLIFTIGGYLLLALAVNTFGVQEEEETR---
MMAR_4743|M.marinum_M DQTWAALG--EHLDDRQRMDYVFTVGCYALLAMAFNTFGVQLEELEQDRA
MSMEG_4839|M.smegmatis_MC2_155 DPTWAALG--EKLDDRQRMDLVFTVGGYGLMAMAYNTFGIEPESGH----
MLBr_02465|M.leprae_Br4923 SSSWQQLE--THLDRRRLIEFCMLISQYDGLAATISSLDIPLDNSC----
MUL_4533|M.ulcerans_Agy99 PATWEQLE--IHLNRRRLIEFCLLATQYDALAATITALGVPLDNPR----
MAB_4119|M.abscessus_ATCC_1997 DEAWEGLS--KFLDRRQLIGFCLLVTQYDGLAATMSSLRIPLDH------
:* : : :. :: : :
Mb1558|M.bovis_AF2122/97 PDGLSPR
Rv1531|M.tuberculosis_H37Rv PDGLSPR
Mflv_2473|M.gilvum_PYR-GCK -------
Mvan_4181|M.vanbaalenii_PYR-1 -------
TH_3878|M.thermoresistible__bu -------
MAV_3958|M.avium_104 -------
MMAR_4743|M.marinum_M ER-----
MSMEG_4839|M.smegmatis_MC2_155 -------
MLBr_02465|M.leprae_Br4923 -------
MUL_4533|M.ulcerans_Agy99 -------
MAB_4119|M.abscessus_ATCC_1997 -------