For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTSRVPLLPVDEAKAAADEAGVPDYMAELSIFQVLLNHPRLARTFNDLLATMLWHGTLDSRLRELVIMR IGWLTDCDYEWTQHWRVASGLGVSADDLLGVRDWQGYNGFGPAEQAVLAATDDVVREGAVSAQSWSACER ELHCDKVVLIELVTVISAWRMVASILHSLEVPLEDGVSSWPPDGLSPR
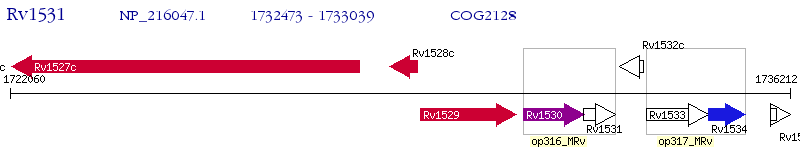
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1531 | - | - | 100% (188) | hypothetical protein Rv1531 |
| M. tuberculosis H37Rv | Rv0464c | - | 3e-12 | 29.52% (166) | hypothetical protein Rv0464c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1558 | - | 1e-109 | 100.00% (188) | hypothetical protein Mb1558 |
| M. gilvum PYR-GCK | Mflv_4627 | - | 1e-57 | 54.44% (180) | carboxymuconolactone decarboxylase |
| M. leprae Br4923 | MLBr_02465 | - | 9e-13 | 32.80% (125) | hypothetical protein MLBr_02465 |
| M. abscessus ATCC 19977 | MAB_2040c | - | 1e-57 | 53.26% (184) | hypothetical protein MAB_2040c |
| M. marinum M | MMAR_2345 | - | 4e-92 | 85.56% (187) | hypothetical protein MMAR_2345 |
| M. avium 104 | MAV_3238 | pcaC | 1e-85 | 79.68% (187) | hypothetical protein MAV_3238 |
| M. smegmatis MC2 155 | MSMEG_6536 | - | 2e-14 | 32.39% (142) | carboxymuconolactone decarboxylase |
| M. thermoresistible (build 8) | TH_2018 | - | 5e-15 | 31.21% (141) | carboxymuconolactone decarboxylase |
| M. ulcerans Agy99 | MUL_4533 | - | 6e-12 | 29.59% (169) | hypothetical protein MUL_4533 |
| M. vanbaalenii PYR-1 | Mvan_4212 | - | 8e-18 | 30.46% (174) | carboxymuconolactone decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4627|M.gilvum_PYR-GCK --------------MLALTDARERAAQCGIPDAMAELS----VFRIALHQ
MAB_2040c|M.abscessus_ATCC_199 ------MTMAERVPMLDREQAQLRAAECGLPEELAELS----VFRVALHQ
Rv1531|M.tuberculosis_H37Rv -------MTTSRVPLLPVDEAKAAADEAGVPDYMAELS----IFQVLLNH
Mb1558|M.bovis_AF2122/97 -------MTTSRVPLLPVDEAKAAADEAGVPDYMAELS----IFQVLLNH
MMAR_2345|M.marinum_M -------MTIPRVPLLPVNEAKAAADEAGVPNYMAELS----IFQALLNH
MAV_3238|M.avium_104 -------MPTARLPKLPLDEAKAAADEAAVPNYMAELS----IFQVLLNH
TH_2018|M.thermoresistible__bu -------VRLAPLPAEQWDDEVLSALAVMLPEERRNPDGAGTALSTLVRH
Mvan_4212|M.vanbaalenii_PYR-1 -------MRLAPLPADQWDGVAEQALSGLMPAELRNPETAGNLVGTLLRH
MSMEG_6536|M.smegmatis_MC2_155 --------MLPPLPAEDWDDAARDALASVVPPERRDADGVGNALGVLVRH
MLBr_02465|M.leprae_Br4923 MSGGTGTGPVGRIPPGSLRQLGPINWVIAKLAASLLRTSEMHLFTILGQR
MUL_4533|M.ulcerans_Agy99 MTG----DQVARIPSGGIRQLGPVNWALAKLAARSVRAPEMHLFTALGYR
. :
Mflv_4627|M.gilvum_PYR-GCK PPVAVALHGMLEALLWKGALDARLRELIIMRIGWVTDSVYEWTQHWRVAR
MAB_2040c|M.abscessus_ATCC_199 PRVAVALYGLLDALLFRGSLDARLRELVILRIGWITGSEYEWTQHWRIAT
Rv1531|M.tuberculosis_H37Rv PRLARTFNDLLATMLWHGTLDSRLRELVIMRIGWLTDCDYEWTQHWRVAS
Mb1558|M.bovis_AF2122/97 PRLARTFNDLLATMLWHGTLDSRLRELVIMRIGWLTDCDYEWTQHWRVAS
MMAR_2345|M.marinum_M PRLARTFNDLLATMLWHGALDPRLRELVIMRIGWLTACDYEWTQHWRVAS
MAV_3238|M.avium_104 PPLARAINDLLATMLWHGALDSRLRELIIMRIGWLTGCDYEWTQHWRVAS
TH_2018|M.thermoresistible__bu PRLTKAFLRFSNHLLFRSTLPPRLRELAVLRVAHRRSCDYEWAHHVFIGK
Mvan_4212|M.vanbaalenii_PYR-1 PKLARAFLRFNFQLLYGTTLPERLREFVVLRVARLSNCEYEWRHHVAMGR
MSMEG_6536|M.smegmatis_MC2_155 PELARHFLQFNNYLQRDSLLPERIRELVILRTAYRAGCVYEWGHHAAMGL
MLBr_02465|M.leprae_Br4923 QLLFWAWLIYGGRLLR-GKLPRVDTELVILRVAHLRTCEYELQHHRRMAR
MUL_4533|M.ulcerans_Agy99 QYLFWTWAIYSGRLLH-GRLPRIDTELVILRVAHLRSCEYELQHHRRMAR
: : * *: ::* . . ** :* :.
Mflv_4627|M.gilvum_PYR-GCK LLDVPERDLLGVRDWQNA-------GHFGEAERAVLAATDETLRDGTISD
MAB_2040c|M.abscessus_ATCC_199 LLGVPERDLLAVRDWQNS-------ESLGPVERAVLAATDDVVRDGVISE
Rv1531|M.tuberculosis_H37Rv GLGVSADDLLGVRDWQGY-------NGFGPAEQAVLAATDDVVREGAVSA
Mb1558|M.bovis_AF2122/97 GLGVSADDLLGVRDWQGY-------NGFGPAEQAVLAATDDVVREGAVSA
MMAR_2345|M.marinum_M GLGVSAQDLLGVRDWRQY-------NGFGPTERAVLAATDDIVRDGAVSA
MAV_3238|M.avium_104 RLGVPGDDLLGVRDWRNY-------PGFGPTERAVLAATDDVVRHGSVSA
TH_2018|M.thermoresistible__bu AEGLTDDDIAGVQRGAVD-------DPF---DQAVLDAVDELEEHYRISD
Mvan_4212|M.vanbaalenii_PYR-1 DAGLSDDVIAGIERGEAV-------DEF---DRAVLTAVDELHHDSVISD
MSMEG_6536|M.smegmatis_MC2_155 LAGLTVDDVESARSGSAA-------DEF---DQTVLDATDELVAGAQLSP
MLBr_02465|M.leprae_Br4923 KRGLDTKIQAMIFAWPDVPTGAG----LSVRQQALLAATDEFVKDRKITS
MUL_4533|M.ulcerans_Agy99 AAGLDAHAQATIFAWPDAPQGDGPRRVLSSRQQALLTATDELIKNRSVTP
.: : ::::* *.*: ::
Mflv_4627|M.gilvum_PYR-GCK ETWAECQRALHGDPAVLVELVAAIGNWRLFSALLRSLHVPLEDGLEGWPP
MAB_2040c|M.abscessus_ATCC_199 ENWAGCQKAFNSDHAVLVELVGAIANWRLFSILLRSLNIPLESGTDGWPP
Rv1531|M.tuberculosis_H37Rv QSWSACERELHCDKVVLIELVTVISAWRMVASILHSLEVPLEDGVSSWPP
Mb1558|M.bovis_AF2122/97 QSWSACERELHCDKVVLIELVTVISAWRMVASILHSLEVPLEDGVSSWPP
MMAR_2345|M.marinum_M SSWAECERELHGDVTVLTELVTAISAWCMVASILHSLKVPLEDGVASWPP
MAV_3238|M.avium_104 ASWAQCEHEFKRDTTVLVELVTVIGAWRMVASILQSLEVPLEDGVASWPP
TH_2018|M.thermoresistible__bu ATWATLGARLD--ERQLMDLVFTVGCYGLMAMAYNTFGITPETGR-----
Mvan_4212|M.vanbaalenii_PYR-1 STWTALSSHLD--ERQLMDLVFTIGCYGALAMAINTFGVEPDQER-----
MSMEG_6536|M.smegmatis_MC2_155 ETWTRLGRWLD--DRQRMDLMFTVGGYLTLAMALNTFEVQLEEGAGRPEF
MLBr_02465|M.leprae_Br4923 SSWQQLETHLD--RRRLIEFCMLISQYDGLAATISSLDIPLDNSC-----
MUL_4533|M.ulcerans_Agy99 ATWEQLEIHLN--RRRLIEFCLLATQYDALAATITALGVPLDNPR-----
.* :. :: : .: :: : :
Mflv_4627|M.gilvum_PYR-GCK DGVRPSVD-
MAB_2040c|M.abscessus_ATCC_199 DGRAPRRRD
Rv1531|M.tuberculosis_H37Rv DGLSPR---
Mb1558|M.bovis_AF2122/97 DGLSPR---
MMAR_2345|M.marinum_M DGCSP----
MAV_3238|M.avium_104 DGREPAS--
TH_2018|M.thermoresistible__bu ---------
Mvan_4212|M.vanbaalenii_PYR-1 ---------
MSMEG_6536|M.smegmatis_MC2_155 YAPVGTAR-
MLBr_02465|M.leprae_Br4923 ---------
MUL_4533|M.ulcerans_Agy99 ---------