For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRVTPLRFRDWPPEMRDAMAALMPPNPRHPAPVTKDRPKAENALGTLAHHPSLARAFCTLQGHLLMGTTL SMRHREMLILRTAAVRGSAYEWTHHNFIAPDGGISTEDVARIAFGPTAPYWSEFEVALLRCVDELIRDGQ MSDATWATLATEFDTQQLLDTMFTVTSYDALMRMLKTCQVHLEDDVRELRAKAGLSVDGDGA*
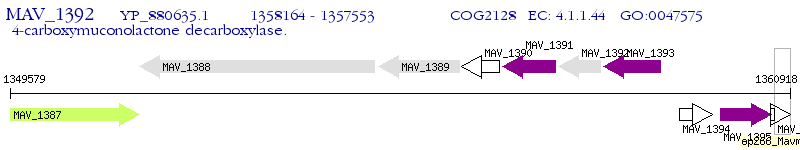
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1392 | pcaC | - | 100% (203) | hypothetical protein MAV_1392 |
| M. avium 104 | MAV_0906 | pcaC | 6e-28 | 39.67% (184) | hypothetical protein MAV_0906 |
| M. avium 104 | MAV_3958 | pcaC | 3e-22 | 33.87% (186) | hypothetical protein MAV_3958 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1558 | - | 1e-13 | 30.20% (149) | hypothetical protein Mb1558 |
| M. gilvum PYR-GCK | Mflv_2440 | - | 3e-25 | 34.78% (184) | carboxymuconolactone decarboxylase |
| M. tuberculosis H37Rv | Rv1531 | - | 1e-13 | 30.20% (149) | hypothetical protein Rv1531 |
| M. leprae Br4923 | MLBr_02465 | - | 9e-12 | 30.16% (126) | hypothetical protein MLBr_02465 |
| M. abscessus ATCC 19977 | MAB_4119 | - | 4e-15 | 37.70% (122) | hypothetical protein MAB_4119 |
| M. marinum M | MMAR_4743 | - | 6e-29 | 38.25% (183) | hypothetical protein MMAR_4743 |
| M. smegmatis MC2 155 | MSMEG_6536 | - | 2e-27 | 38.33% (180) | carboxymuconolactone decarboxylase |
| M. thermoresistible (build 8) | TH_2018 | - | 7e-28 | 39.88% (173) | carboxymuconolactone decarboxylase |
| M. ulcerans Agy99 | MUL_4533 | - | 1e-10 | 30.34% (145) | hypothetical protein MUL_4533 |
| M. vanbaalenii PYR-1 | Mvan_4212 | - | 7e-29 | 38.04% (184) | carboxymuconolactone decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1558|M.bovis_AF2122/97 ---------MTTSRVPLLPVDEAKAAADEAGVPDYMAELS----------
Rv1531|M.tuberculosis_H37Rv ---------MTTSRVPLLPVDEAKAAADEAGVPDYMAELS----------
MMAR_4743|M.marinum_M ---------MRLPPLPADQWDEAVQRSLSVILPAQRCNPR-------DAG
TH_2018|M.thermoresistible__bu ---------VRLAPLPAEQWDDEVLSALAVMLPEERRNPD-------GAG
Mflv_2440|M.gilvum_PYR-GCK ---------MRVAPLPADQWDDTVDKALSGLMPEEMRNPE-------TAG
Mvan_4212|M.vanbaalenii_PYR-1 ---------MRLAPLPADQWDGVAEQALSGLMPAELRNPE-------TAG
MSMEG_6536|M.smegmatis_MC2_155 ----------MLPPLPAEDWDDAARDALASVVPPERRDAD-------GVG
MAV_1392|M.avium_104 --------MSRVTPLRFRDWPPEMRDAMAALMPPNPRHPAPVTKDRPKAE
MLBr_02465|M.leprae_Br4923 MSGGTGTGPVGRIPPGSLRQLGPINWVIAKLAASLLRTSE---------M
MUL_4533|M.ulcerans_Agy99 MTG----DQVARIPSGGIRQLGPVNWALAKLAARSVRAPE---------M
MAB_4119|M.abscessus_ATCC_1997 MSS-------GRIPSGGLRELGPINWVIAKGMARAVNAPE---------M
.
Mb1558|M.bovis_AF2122/97 -IFQVLLNHPRLARTFNDLLATMLWHGTLDSRLRELVIMRIGWLTDCDYE
Rv1531|M.tuberculosis_H37Rv -IFQVLLNHPRLARTFNDLLATMLWHGTLDSRLRELVIMRIGWLTDCDYE
MMAR_4743|M.marinum_M NALSTFANHPALAKAFLRFNTHLLFASTLPARVRELAILRVAHRRACAYE
TH_2018|M.thermoresistible__bu TALSTLVRHPRLTKAFLRFSNHLLFRSTLPPRLRELAVLRVAHRRSCDYE
Mflv_2440|M.gilvum_PYR-GCK NLVATLVRHPRLAKGFLRFNFHLLYGSSLPARLRELAVLRIAHLTKCEYE
Mvan_4212|M.vanbaalenii_PYR-1 NLVGTLLRHPKLARAFLRFNFQLLYGTTLPERLREFVVLRVARLSNCEYE
MSMEG_6536|M.smegmatis_MC2_155 NALGVLVRHPELARHFLQFNNYLQRDSLLPERIRELVILRTAYRAGCVYE
MAV_1392|M.avium_104 NALGTLAHHPSLARAFCTLQGHLLMGTTLSMRHREMLILRTAAVRGSAYE
MLBr_02465|M.leprae_Br4923 HLFTILGQRQLLFWAWLIYGGRLLRG-KLPRVDTELVILRVAHLRTCEYE
MUL_4533|M.ulcerans_Agy99 HLFTALGYRQYLFWTWAIYSGRLLHG-RLPRIDTELVILRVAHLRSCEYE
MAB_4119|M.abscessus_ATCC_1997 HLATTLGQTGARFWPWLAYSGAILRGTKLSTRDTEVVILRVAHVRECEYE
: : : * *. ::* . . **
Mb1558|M.bovis_AF2122/97 WTQHWRVASGLGVSADDLLGVRDW---QGYNG----FGPAEQAVLAATDD
Rv1531|M.tuberculosis_H37Rv WTQHWRVASGLGVSADDLLGVRDW---QGYNG----FGPAEQAVLAATDD
MMAR_4743|M.marinum_M WTHHVALAEQAGISGDEIAAVRRG---EAT-------DDFERAVLTGVDE
TH_2018|M.thermoresistible__bu WAHHVFIGKAEGLTDDDIAGVQRG---AVD-------DPFDQAVLDAVDE
Mflv_2440|M.gilvum_PYR-GCK WLHHVEMGREAGLSDDVIDGITRG---EAT-------DGLDRAVLNAVDE
Mvan_4212|M.vanbaalenii_PYR-1 WRHHVAMGRDAGLSDDVIAGIERG---EAV-------DEFDRAVLTAVDE
MSMEG_6536|M.smegmatis_MC2_155 WGHHAAMGLLAGLTVDDVESARSG---SAA-------DEFDQTVLDATDE
MAV_1392|M.avium_104 WTHHNFIAPDGGISTEDVARIAFG---PTAPY----WSEFEVALLRCVDE
MLBr_02465|M.leprae_Br4923 LQHHRRMARKRGLDTKIQAMIFAWPDVPTGAG----LSVRQQALLAATDE
MUL_4533|M.ulcerans_Agy99 LQHHRRMARAAGLDAHAQATIFAWPDAPQGDGPRRVLSSRQQALLTATDE
MAB_4119|M.abscessus_ATCC_1997 LQHHTRIAKSAGIDPAYQERIFTG---AGAEG----LSDKERALITGVDE
:* :. *: . : ::: .*:
Mb1558|M.bovis_AF2122/97 VVREGAVSAQSWSACERELHCDKVVLIELVTVISAWRMVASILHSLEVPL
Rv1531|M.tuberculosis_H37Rv VVREGAVSAQSWSACERELHCDKVVLIELVTVISAWRMVASILHSLEVPL
MMAR_4743|M.marinum_M LDENSELSDQTWAALG--EHLDDRQRMDYVFTVGCYALLAMAFNTFGVQL
TH_2018|M.thermoresistible__bu LEEHYRISDATWATLG--ARLDERQLMDLVFTVGCYGLMAMAYNTFGITP
Mflv_2440|M.gilvum_PYR-GCK LQNDSVVSDATWTALS--DHLDERQLMDLVFTIGCYGALAMAINTFGVEP
Mvan_4212|M.vanbaalenii_PYR-1 LHHDSVISDSTWTALS--SHLDERQLMDLVFTIGCYGALAMAINTFGVEP
MSMEG_6536|M.smegmatis_MC2_155 LVAGAQLSPETWTRLG--RWLDDRQRMDLMFTVGGYLTLAMALNTFEVQL
MAV_1392|M.avium_104 LIRDGQMSDATWATLA--TEFDTQQLLDTMFTVTSYDALMRMLKTCQVHL
MLBr_02465|M.leprae_Br4923 FVKDRKITSSSWQQLE--THLDRRRLIEFCMLISQYDGLAATISSLDIPL
MUL_4533|M.ulcerans_Agy99 LIKNRSVTPATWEQLE--IHLNRRRLIEFCLLATQYDALAATITALGVPL
MAB_4119|M.abscessus_ATCC_1997 ILTTRTLSDEAWEGLS--KFLDRRQLIGFCLLVTQYDGLAATMSSLRIPL
. :: :* : : : : : :
Mb1558|M.bovis_AF2122/97 EDGVSSWPPDGLSPR-----
Rv1531|M.tuberculosis_H37Rv EDGVSSWPPDGLSPR-----
MMAR_4743|M.marinum_M EELEQDRAER----------
TH_2018|M.thermoresistible__bu ETGR----------------
Mflv_2440|M.gilvum_PYR-GCK DQER----------------
Mvan_4212|M.vanbaalenii_PYR-1 DQER----------------
MSMEG_6536|M.smegmatis_MC2_155 EEGAGRPEFYAPVGTAR---
MAV_1392|M.avium_104 EDDVRELRAKAGLSVDGDGA
MLBr_02465|M.leprae_Br4923 DNSC----------------
MUL_4533|M.ulcerans_Agy99 DNPR----------------
MAB_4119|M.abscessus_ATCC_1997 DH------------------
: