For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRLPPLPADQWDEAVQRSLSVILPAQRCNPRDAGNALSTFANHPALAKAFLRFNTHLLFASTLPARVREL AILRVAHRRACAYEWTHHVALAEQAGISGDEIAAVRRGEATDDFERAVLTGVDELDENSELSDQTWAALG EHLDDRQRMDYVFTVGCYALLAMAFNTFGVQLEELEQDRAER
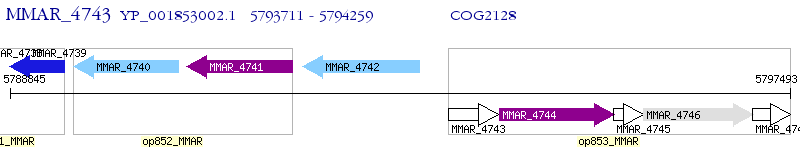
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4743 | - | - | 100% (182) | hypothetical protein MMAR_4743 |
| M. marinum M | MMAR_2345 | - | 3e-14 | 31.16% (138) | hypothetical protein MMAR_2345 |
| M. marinum M | MMAR_3164 | - | 5e-13 | 28.89% (180) | hypothetical protein MMAR_3164 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1558 | - | 1e-16 | 30.39% (181) | hypothetical protein Mb1558 |
| M. gilvum PYR-GCK | Mflv_2440 | - | 1e-58 | 57.47% (174) | carboxymuconolactone decarboxylase |
| M. tuberculosis H37Rv | Rv1531 | - | 1e-16 | 30.39% (181) | hypothetical protein Rv1531 |
| M. leprae Br4923 | MLBr_02465 | - | 7e-11 | 30.08% (123) | hypothetical protein MLBr_02465 |
| M. abscessus ATCC 19977 | MAB_4119 | - | 6e-16 | 36.22% (127) | hypothetical protein MAB_4119 |
| M. avium 104 | MAV_0906 | pcaC | 1e-66 | 69.78% (182) | hypothetical protein MAV_0906 |
| M. smegmatis MC2 155 | MSMEG_4839 | - | 6e-59 | 60.69% (173) | carboxymuconolactone decarboxylase |
| M. thermoresistible (build 8) | TH_2018 | - | 6e-60 | 60.69% (173) | carboxymuconolactone decarboxylase |
| M. ulcerans Agy99 | MUL_4533 | - | 8e-12 | 32.54% (126) | hypothetical protein MUL_4533 |
| M. vanbaalenii PYR-1 | Mvan_4212 | - | 3e-57 | 57.47% (174) | carboxymuconolactone decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1558|M.bovis_AF2122/97 -------MTTSRVPLLPVDEAKAAADEAGVPD----YMAELSIFQVLLNH
Rv1531|M.tuberculosis_H37Rv -------MTTSRVPLLPVDEAKAAADEAGVPD----YMAELSIFQVLLNH
MMAR_4743|M.marinum_M -------MRLPPLPADQWDEAVQRSLSVILPAQRCNPRDAGNALSTFANH
MAV_0906|M.avium_104 -------MRLQPLPAEQWDEATRQALAAMRGA------DTNNALSTLAHH
MSMEG_4839|M.smegmatis_MC2_155 -------MRLTPLPADEWDDEVRLALSVMLPEERLNPEGAGTALSTLARH
TH_2018|M.thermoresistible__bu -------VRLAPLPAEQWDDEVLSALAVMLPEERRNPDGAGTALSTLVRH
Mflv_2440|M.gilvum_PYR-GCK -------MRVAPLPADQWDDTVDKALSGLMPEEMRNPETAGNLVATLVRH
Mvan_4212|M.vanbaalenii_PYR-1 -------MRLAPLPADQWDGVAEQALSGLMPAELRNPETAGNLVGTLLRH
MLBr_02465|M.leprae_Br4923 MSGGTGTGPVGRIPPGSLRQLGPINWVIAKLAASLLRTSEMHLFTILGQR
MUL_4533|M.ulcerans_Agy99 MTG----DQVARIPSGGIRQLGPVNWALAKLAARSVRAPEMHLFTALGYR
MAB_4119|M.abscessus_ATCC_1997 MSS-------GRIPSGGLRELGPINWVIAKGMARAVNAPEMHLATTLGQT
:* :
Mb1558|M.bovis_AF2122/97 PRLARTFNDLLATMLWHGTLDSRLRELVIMRIGWLTDCDYEWTQHWRVAS
Rv1531|M.tuberculosis_H37Rv PRLARTFNDLLATMLWHGTLDSRLRELVIMRIGWLTDCDYEWTQHWRVAS
MMAR_4743|M.marinum_M PALAKAFLRFNTHLLFASTLPARVRELAILRVAHRRACAYEWTHHVALAE
MAV_0906|M.avium_104 PALAKAFLRFNVHLLTASTLPPRVRELAILRVAHRRQCAYEWSHHVGMAK
MSMEG_4839|M.smegmatis_MC2_155 PRLTKAFLRFSNHLLFRSTLDPRLRELAILRIARRRNCEYEWAHHAFIGK
TH_2018|M.thermoresistible__bu PRLTKAFLRFSNHLLFRSTLPPRLRELAVLRVAHRRSCDYEWAHHVFIGK
Mflv_2440|M.gilvum_PYR-GCK PRLAKGFLRFNFHLLYGSSLPARLRELAVLRIAHLTKCEYEWLHHVEMGR
Mvan_4212|M.vanbaalenii_PYR-1 PKLARAFLRFNFQLLYGTTLPERLREFVVLRVARLSNCEYEWRHHVAMGR
MLBr_02465|M.leprae_Br4923 QLLFWAWLIYGGRLLRG-KLPRVDTELVILRVAHLRTCEYELQHHRRMAR
MUL_4533|M.ulcerans_Agy99 QYLFWTWAIYSGRLLHG-RLPRIDTELVILRVAHLRSCEYELQHHRRMAR
MAB_4119|M.abscessus_ATCC_1997 GARFWPWLAYSGAILRGTKLSTRDTEVVILRVAHVRECEYELQHHTRIAK
: :* * *..::*:. * ** :* :.
Mb1558|M.bovis_AF2122/97 GLGVSADDLLGVRDW----QGYNG---FGPAEQAVLAATDDVVREGAVSA
Rv1531|M.tuberculosis_H37Rv GLGVSADDLLGVRDW----QGYNG---FGPAEQAVLAATDDVVREGAVSA
MMAR_4743|M.marinum_M QAGISGDEIAAVR------RGEAT----DDFERAVLTGVDELDENSELSD
MAV_0906|M.avium_104 DEGITDEQIAAVRCF----AGDGAGP-FDAFDHAVLAGVDELDEKSELSD
MSMEG_4839|M.smegmatis_MC2_155 AEGLTDDDIDGIG------RGVVS----DPFDQVVIDAVDELDELSTISD
TH_2018|M.thermoresistible__bu AEGLTDDDIAGVQ------RGAVD----DPFDQAVLDAVDELEEHYRISD
Mflv_2440|M.gilvum_PYR-GCK EAGLSDDVIDGIT------RGEAT----DGLDRAVLNAVDELQNDSVVSD
Mvan_4212|M.vanbaalenii_PYR-1 DAGLSDDVIAGIE------RGEAV----DEFDRAVLTAVDELHHDSVISD
MLBr_02465|M.leprae_Br4923 KRGLDTKIQAMIFAWPDVPTGAG----LSVRQQALLAATDEFVKDRKITS
MUL_4533|M.ulcerans_Agy99 AAGLDAHAQATIFAWPDAPQGDGPRRVLSSRQQALLTATDELIKNRSVTP
MAB_4119|M.abscessus_ATCC_1997 SAGIDPAYQERIFTG---AGAEG----LSDKERALITGVDEILTTRTLSD
*: : . . ::.:: ..*:. ::
Mb1558|M.bovis_AF2122/97 QSWSACERELHCDKVVLIELVTVISAWRMVASILHSLEVPLEDGVSSWPP
Rv1531|M.tuberculosis_H37Rv QSWSACERELHCDKVVLIELVTVISAWRMVASILHSLEVPLEDGVSSWPP
MMAR_4743|M.marinum_M QTWAALG--EHLDDRQRMDYVFTVGCYALLAMAFNTFGVQLEELEQDRAE
MAV_0906|M.avium_104 RTWAALG--ERLDDRQRMDYVFTVGCYTLLAMAFNTFGIQLEHAEQH---
MSMEG_4839|M.smegmatis_MC2_155 PTWAALG--EKLDDRQRMDLVFTVGGYGLMAMAYNTFGIEPESGH-----
TH_2018|M.thermoresistible__bu ATWATLG--ARLDERQLMDLVFTVGCYGLMAMAYNTFGITPETGR-----
Mflv_2440|M.gilvum_PYR-GCK ATWTALS--DHLDERQLMDLVFTIGCYGALAMAINTFGVEPDQER-----
Mvan_4212|M.vanbaalenii_PYR-1 STWTALS--SHLDERQLMDLVFTIGCYGALAMAINTFGVEPDQER-----
MLBr_02465|M.leprae_Br4923 SSWQQLE--THLDRRRLIEFCMLISQYDGLAATISSLDIPLDNSC-----
MUL_4533|M.ulcerans_Agy99 ATWEQLE--IHLNRRRLIEFCLLATQYDALAATITALGVPLDNPR-----
MAB_4119|M.abscessus_ATCC_1997 EAWEGLS--KFLDRRQLIGFCLLVTQYDGLAATMSSLRIPLDH-------
:* : : : :* :: : :
Mb1558|M.bovis_AF2122/97 DGLSPR
Rv1531|M.tuberculosis_H37Rv DGLSPR
MMAR_4743|M.marinum_M R-----
MAV_0906|M.avium_104 ------
MSMEG_4839|M.smegmatis_MC2_155 ------
TH_2018|M.thermoresistible__bu ------
Mflv_2440|M.gilvum_PYR-GCK ------
Mvan_4212|M.vanbaalenii_PYR-1 ------
MLBr_02465|M.leprae_Br4923 ------
MUL_4533|M.ulcerans_Agy99 ------
MAB_4119|M.abscessus_ATCC_1997 ------