For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSASCHGLARVAVFAVAGATAVSLSACGTSNKSSPTSTSTSTSTVTSTSTSAAPNAEAKVSGLIASVAGN SIQVTKEDNGTAAVNFTSTTKITEAVPAGLPDVTPGSCVSVKPTEGSAPGQPVTAASVKISQSVNGACPK PHQPTPGGSSSPAPSLPPSQAPAKPALVRGSVASVSGNTINVTGTDPSGNTTQTTVTVDDKTKYTKQSSA NTDAITPGKCLSARGTNDSGGALQATSIQLRQAVDGKCGKPKQPGHGG
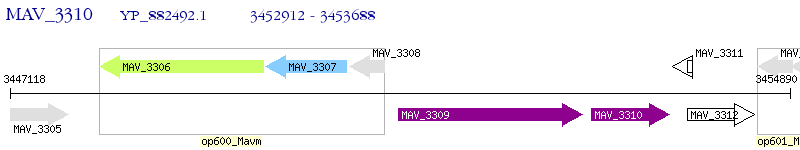
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3310 | - | - | 100% (258) | 27 kDa lipoprotein antigen |
| M. avium 104 | MAV_3312 | - | 3e-30 | 38.05% (226) | hypothetical protein MAV_3312 |
| M. avium 104 | MAV_4561 | - | 7e-06 | 29.87% (154) | LpqN protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1824 | - | 1e-06 | 29.20% (137) | Pro-rich protease |
| M. gilvum PYR-GCK | Mflv_0760 | - | 1e-05 | 25.13% (195) | hypothetical protein Mflv_0760 |
| M. tuberculosis H37Rv | Rv1796 | mycP5 | 1e-06 | 29.20% (137) | proline rich membrane-anchored mycosin MYCP5 (serine |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2743 | - | 3e-50 | 45.78% (249) | hypothetical protein MAB_2743 |
| M. marinum M | MMAR_0950 | lpqN | 2e-07 | 34.03% (144) | lipoprotein, LpqN |
| M. smegmatis MC2 155 | MSMEG_1600 | - | 2e-06 | 24.00% (250) | hypothetical protein MSMEG_1600 |
| M. thermoresistible (build 8) | TH_2316 | - | 2e-06 | 24.02% (204) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0701 | lpqN | 4e-07 | 33.80% (142) | lipoprotein, LpqN |
| M. vanbaalenii PYR-1 | Mvan_1509 | - | 3e-06 | 27.27% (154) | hypothetical protein Mvan_1509 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3310|M.avium_104 ------------------------------MSASCHGLARVAVFAVAGAT
MAB_2743|M.abscessus_ATCC_1997 --------------------------MTPTQTLAQPRFARGALLAFAGLT
Mb1824|M.bovis_AF2122/97 -----------------------MQRFGTGSSRSWCGRAGTATIAAVLLA
Rv1796|M.tuberculosis_H37Rv -----------------------MQRFGTGSSRSWCGRAGTATIAAVLLA
MSMEG_1600|M.smegmatis_MC2_155 -------------------MPDFGRWTSNGGDPSLNEINRTDRFIEALSL
Mvan_1509|M.vanbaalenii_PYR-1 -------------------MPDHGRWNS-GGDPSLNEINRTDQFLDALAS
MMAR_0950|M.marinum_M -------------------------------MKDLTAVLATAALSLTLVA
MUL_0701|M.ulcerans_Agy99 -------------------------------MKDLTAVLATAALSLTLVA
Mflv_0760|M.gilvum_PYR-GCK -------------------MFVDGVWTGGAADAAKSAVENRLQQMHSLIA
TH_2316|M.thermoresistible__bu VGRVGALAVALGVGVAVATGGTAVAWADSGSTASSSSSQDSSSKSNPAQT
MAV_3310|M.avium_104 AVS--------------------LSACGTSNKSSPTSTSTSTSTVTSTST
MAB_2743|M.abscessus_ATCC_1997 AFS--------------------VAACGTHGDTNAASTRSSAPTTSSSAP
Mb1824|M.bovis_AF2122/97 SGA--------------------LTGLPPAYAISPPTIDPGALPPDGPPG
Rv1796|M.tuberculosis_H37Rv SGA--------------------LTGLPPAYAISPPTIDPGALPPDGPPG
MSMEG_1600|M.smegmatis_MC2_155 EQP--------------------VYSTDPEEAELAFLLAGWRDDVRQAPM
Mvan_1509|M.vanbaalenii_PYR-1 QQP--------------------VYATDRGEAELAHLLAGWRDDVRETPM
MMAR_0950|M.marinum_M CGS--------------------GTTSDNKTSTSTSTSTSTSTSTTTSAT
MUL_0701|M.ulcerans_Agy99 CGS--------------------GTTSDNK--TSTSTSTSTSTSTTTSAT
Mflv_0760|M.gilvum_PYR-GCK QLEKAIAWYS--------MIARVVVDAKLAVNDNLLAAQDYFSVIRKSPE
TH_2316|M.thermoresistible__bu RSSAKSGIGSGAGSGPGSRLDRPASRAPLGGSGADSSATEQTGAVSSGTS
.
MAV_3310|M.avium_104 SAAPNAEAKVSGLIASVAGNSIQVTKED----NGTAAVNFTSTTKITEAV
MAB_2743|M.abscessus_ATCC_1997 HGEHNEKGHVSGLIVSVSGNSVEVTKQ-----NGNATVSFGQSTKVSEIF
Mb1824|M.bovis_AF2122/97 PLAPMKQNAYCTEVGVLPGTDFQLQPKYMEMLNLNEAWQFGRGDGVKVAV
Rv1796|M.tuberculosis_H37Rv PLAPMKQNAYCTEVGVLPGTDFQLQPKYMEMLNLNEAWQFGRGDGVKVAV
MSMEG_1600|M.smegmatis_MC2_155 TGIINPREAVVALDRAVARNRPRLPLALVGSAAAAVLCLGGFGAAVYGSS
Mvan_1509|M.vanbaalenii_PYR-1 GHVMTAEDAAIALGRVTKPPRRRLSLAVIGSAAAAVLAIGGVGAVVAGSG
MMAR_0950|M.marinum_M SPTPGAQANYTIADYIRDNDIQETPVHHGDPGSPNIDLPVPDG-------
MUL_0701|M.ulcerans_Agy99 SPTPGAQANYTIADYIRDNDIQETPVHHGDPGSPNIDLPVPDG-------
Mflv_0760|M.gilvum_PYR-GCK LEPGEKEALIQNWVRSQNVVNTAIILGAMAKVPAFTTWQPPPQPVRVPHS
TH_2316|M.thermoresistible__bu AWRSAGMPAGTSLTGRLADSVRDAVKPWRPATTRSTDPTPVETPAEANPE
MAV_3310|M.avium_104 PAG-------LPDVTPG----------------------SCVSVKPTEGS
MAB_2743|M.abscessus_ATCC_1997 AAQ-------LSDVTAG----------------------SCIAAMTPPDS
Mb1824|M.bovis_AF2122/97 IDTGVTPHPRLPRLIPGGDYVMAGGDGLSDCDAHGTLVASMIAAVPANGA
Rv1796|M.tuberculosis_H37Rv IDTGVTPHPRLPRLIPGGDYVMAGGDGLSDCDAHGTLVASMIAAVPANGA
MSMEG_1600|M.smegmatis_MC2_155 PGDALYG---LRGMIFG----EQATR--DVQVELASTELEQVQQLIEQGD
Mvan_1509|M.vanbaalenii_PYR-1 PGDALYG---LRTMLFG----EQAQTRDDAVILAAQTEMQQVQQLIEQGD
MMAR_0950|M.marinum_M ------------------------------------------WQLLPEGS
MUL_0701|M.ulcerans_Agy99 ------------------------------------------WQLLPEGS
Mflv_0760|M.gilvum_PYR-GCK PDTSAASVPKSSLDHFGPVRLTSPRVVLASNRVGPKEAPELAANANPHGP
TH_2316|M.thermoresistible__bu AQPDAHAGSPGESTAPETQPAPSSRGIRSMTAVLPDTESTNTESTNTEST
.
MAV_3310|M.avium_104 APGQPVTAASVKISQSVNG-------------------ACPKPHQPTPGG
MAB_2743|M.abscessus_ATCC_1997 NP---PTARRVMMWPSADG-------------------SCVREH-PKEQQ
Mb1824|M.bovis_AF2122/97 VPLPSVPRRPVTIPTTETPPPPQTVTLSPVPPQTVTVIPAPPPEEGVPPG
Rv1796|M.tuberculosis_H37Rv VPLPSVPRRPVTIPTTETPPPPQTVTLSPVPPQTVTVIPAPPPEEGVPPG
MSMEG_1600|M.smegmatis_MC2_155 WQAAQQKLQTLTTTVATVNDEAQKQQLVTQWQQLSVKVENQDPNATVAPD
Mvan_1509|M.vanbaalenii_PYR-1 WAGAQAKLEAVTTTVATVADVERKQELVTQWQELTVKVEAQDPVATVPPG
MMAR_0950|M.marinum_M GAPYGGIAYTQPADPSDPP-------------------TIVAILSKLTGN
MUL_0701|M.ulcerans_Agy99 GAPYGGIAYTQPADPSDPP-------------------TIVAILSKLTGN
Mflv_0760|M.gilvum_PYR-GCK RSAVTPPHPATATSPTTSSGGVSTANSPSAASASTPSASSPMSSSGGPPS
TH_2316|M.thermoresistible__bu VQHDDSERRSSLRSTAASVPELSTPVRLADATAVRALAERLTERRSVTLE
:
MAV_3310|M.avium_104 S--SSPAPSLPPSQAPAKPALVRG----SVASVSG---------------
MAB_2743|M.abscessus_ATCC_1997 S--TSPAP---TSGKRPEHEGIRG----TVASVSG---------------
Mb1824|M.bovis_AF2122/97 A--PVPGPEPPPAPGPQPPAVDRGGGTVTVPSYSGGRKIAPIDNPRNPHP
Rv1796|M.tuberculosis_H37Rv A--PVPGPEPPPAPGPQPPAVDRGGGTVTVPSYSGGRKIAPIDNPRNPHP
MSMEG_1600|M.smegmatis_MC2_155 A--PPPVFPEVTITPPAPTDTSAVETTPGSATPSEP--------------
Mvan_1509|M.vanbaalenii_PYR-1 S--PLPTFPDVPVAVLDPGTSSETTSTAPSSETTSP--------------
MMAR_0950|M.marinum_M V--EAAKILEFAPGELKNLPGYEG--------------------------
MUL_0701|M.ulcerans_Agy99 V--EAAKILEFAPGELKNLPGYEG--------------------------
Mflv_0760|M.gilvum_PYR-GCK TGGASSGSPSASTSPASGSPSTASPTTSGAAGAQPSNASPAGAAKAQPSP
TH_2316|M.thermoresistible__bu SVRAFTDVAATTALTSAASVAAATVEPVALITAAPEATPAVDAEQPAAPR
. .
MAV_3310|M.avium_104 ---------------------NTINVTG--------------TDPSGNTT
MAB_2743|M.abscessus_ATCC_1997 ---------------------NAITVTG--------------TDPNGTAT
Mb1824|M.bovis_AF2122/97 SAPSPALGPPPDAFSGIAPGVEIISIRQSSQAFGLKDPYTGDEDPQTAQK
Rv1796|M.tuberculosis_H37Rv SAPSPALGPPPDAFSGIAPGVEIISIRQSSQAFGLKDPYTGDEDPQTAQK
MSMEG_1600|M.smegmatis_MC2_155 --------------------TSPGESATPSDTSDTSEPSSETTVPSSPTP
Mvan_1509|M.vanbaalenii_PYR-1 --------------------TSPSETTSPTSP-------SETTSPTSPSE
MMAR_0950|M.marinum_M ------------------------------------------SNNGSAST
MUL_0701|M.ulcerans_Agy99 ------------------------------------------SNNGSAST
Mflv_0760|M.gilvum_PYR-GCK IQQVFNQSAPLASSAPAQSPAAPPSSAPAPTTPAGAAPTAGTGAGGGLST
TH_2316|M.thermoresistible__bu LVTGLLAAVGLAPFTANSPLAPTQSTPLLALLGWARRETDRAYTQQARSL
MAV_3310|M.avium_104 -QTTVTVDDKTKYTKQSSANTDAITPGKCLSA-----RGTNDS---GGAL
MAB_2743|M.abscessus_ATCC_1997 GPTTVTVNNDTTYAKRAPVDQNAIATGKCMAA-----HGTDDA---SGNL
Mb1824|M.bovis_AF2122/97 IDNVETMARAIVHAANMGASVINISDVMCMSA-----RNVIDQRALGAAV
Rv1796|M.tuberculosis_H37Rv IDNVETMARAIVHAANMGASVINISDVMCMSA-----RNVIDQRALGAAV
MSMEG_1600|M.smegmatis_MC2_155 STAPSSSVPTATSQTPTSTQVPTSTPTSSAAV-----QLPSPVEEEEPAE
Mvan_1509|M.vanbaalenii_PYR-1 TTSPAPSSETTSAPTSSPSPTPSTAPTTTSVG-----AAPP--SSSAPAT
MMAR_0950|M.marinum_M LSGFQAWQLGGSYTKNGKQRAVAQKTVVIPSQ-----DGLFVLQLDADAL
MUL_0701|M.ulcerans_Agy99 LSGFQAWQLGGSYTKNGKQRAVAQKTVVIPSQ-----DGLFVLQLDADAL
Mflv_0760|M.gilvum_PYR-GCK SGGPAPVAGAPAGAAPPAAPPVPLAPPTSPAP-----PAPPPTGAPGAAP
TH_2316|M.thermoresistible__bu VTARPAVVSTAAVVDPLPADLIGETERIGRITGPGITDGPNRLNIYGTDL
.
MAV_3310|M.avium_104 Q--------ATSIQLRQAVDGKCGKPKQPGHGG-----------------
MAB_2743|M.abscessus_ATCC_1997 Q--------ATSITLRAARDGSCDGGGHEGHHQR----------------
Mb1824|M.bovis_AF2122/97 H--------YAAVDKDAVIVAAAGDGSKKDCKQNPIFDPLQPDDPRAWNA
Rv1796|M.tuberculosis_H37Rv H--------YAAVDKDAVIVAAAGDGSKKDCKQNPIFDPLQPDDPRAWNA
MSMEG_1600|M.smegmatis_MC2_155 P--------QPTSVPQAPTSAPAVPAEPSGPSGDPARPPVTATTTVTLPT
Mvan_1509|M.vanbaalenii_PYR-1 T--------TPTST--TATRAPSATSTTVVQSATTTSAPLSTSSSVAAPP
MMAR_0950|M.marinum_M D--------SEQDVLMDATGVIDDQTKITP--------------------
MUL_0701|M.ulcerans_Agy99 D--------SEQDVLMDATGVIDDQTKITP--------------------
Mflv_0760|M.gilvum_PYR-GCK AGLGAGVAPMSATSSSSGAAAAPIPVSQARAERDAVLAASTAGALRRKNN
TH_2316|M.thermoresistible__bu GIMWDMGEWRGQRAVHVVFGDTFSGPNMSGDWISNAIMISTSRTLYENPA
MAV_3310|M.avium_104 --------------------------------------------------
MAB_2743|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1824|M.bovis_AF2122/97 VTTVVTPSWFHDYVLTVGAVDANGQPLSKMSIAGPWVSISAPGTDVVGLS
Rv1796|M.tuberculosis_H37Rv VTTVVTPSWFHDYVLTVGAVDANGQPLSKMSIAGPWVSISAPGTDVVGLS
MSMEG_1600|M.smegmatis_MC2_155 QAGPAQGPNGRGNGPGNGAKPAEPAQPKAPEQPAVQIPL-LPGMGGN---
Mvan_1509|M.vanbaalenii_PYR-1 TPTPSPSPTPTPAPTVDEESVPSPTAPALPTTTTPIILLPVPGEAG----
MMAR_0950|M.marinum_M --------------------------------------------------
MUL_0701|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0760|M.gilvum_PYR-GCK GSDPLQRARHIGAALNVGVKDFGFFWVTAVTTDGTIVVANSYGIGYIPDG
TH_2316|M.thermoresistible__bu GFLEQTGPAFQFIPGGQLLSWFFPAEVTVIPTAGVHANGTQYVNYMSVRQ
MAV_3310|M.avium_104 --------------------------------------------------
MAB_2743|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1824|M.bovis_AF2122/97 PRDDGLINAIDGPDNSLLVPAGTSFSAAIVSGVAALVRAKFPELSAYQII
Rv1796|M.tuberculosis_H37Rv PRDDGLINAIDGPDNSLLVPAGTSFSAAIVSGVAALVRAKFPELSAYQII
MSMEG_1600|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1509|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0950|M.marinum_M --------------------------------------------------
MUL_0701|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0760|M.gilvum_PYR-GCK VRLPENVRMASADDTIPADERAKWATYPILAVQGWAQHHELQLRAVVATE
TH_2316|M.thermoresistible__bu WGVPGSWTTNFSAISRYDQATDTWVTVPSSVRSAGWLRSSTPYRAGDQNF
MAV_3310|M.avium_104 --------------------------------------------------
MAB_2743|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1824|M.bovis_AF2122/97 NRLIHTARPPARG------------------------------VDNQVGY
Rv1796|M.tuberculosis_H37Rv NRLIHTARPPARG------------------------------VDNQVGY
MSMEG_1600|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1509|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0950|M.marinum_M --------------------------------------------------
MUL_0701|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0760|M.gilvum_PYR-GCK EQFANFDPGVAKI------------------------------ILTAEDI
TH_2316|M.thermoresistible__bu QQMAYVLQPVDQVGEDGVRYVYAFGTPSGRQGSVHLSRVPEEHITDVSKY
MAV_3310|M.avium_104 --------------------------------------------------
MAB_2743|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1824|M.bovis_AF2122/97 GVVDPVAALTWDVPKGPAEPPKQLSAPLVVPQPPAPRDMVPIWVAAGGLA
Rv1796|M.tuberculosis_H37Rv GVVDPVAALTWDVPKGPAEPPKQLSAPLVVPQPPAPRDMVPIWVAAGGLA
MSMEG_1600|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1509|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0950|M.marinum_M --------------------------------------------------
MUL_0701|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0760|M.gilvum_PYR-GCK PDDGTMSGRTRLEVISPTAAQQLSAVGDFGLNDLLPPAPTDLEPPADQTA
TH_2316|M.thermoresistible__bu EYWDGNAWVSSAAHAAPIIGESTRSAGLFGFIIDWANDPNVFGGYLGGLF
MAV_3310|M.avium_104 --------------------------------------------------
MAB_2743|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1824|M.bovis_AF2122/97 GALLIGGAVFGTATLMRRSRKQQ---------------------------
Rv1796|M.tuberculosis_H37Rv GALLIGGAVFGTATLMRRSRKQQ---------------------------
MSMEG_1600|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1509|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0950|M.marinum_M --------------------------------------------------
MUL_0701|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0760|M.gilvum_PYR-GCK TLWFEVAKPLMSTNPQRSAAHLQAFITYANHAQELAVYRAHTAAEPADQR
TH_2316|M.thermoresistible__bu GAKTGGNVGEMSVQYNEYLGKYVILYGNGRTNNIEMRIADSPEGPWSDPI
MAV_3310|M.avium_104 --------------------------------------------------
MAB_2743|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1824|M.bovis_AF2122/97 --------------------------------------------------
Rv1796|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_1600|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1509|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0950|M.marinum_M --------------------------------------------------
MUL_0701|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0760|M.gilvum_PYR-GCK LAIADWVYWQHLAVIMNDAVNADASA------------------------
TH_2316|M.thermoresistible__bu ILATPQQYPGLYAPMIHPWSGTGYLLDENGDPDPESLYWNMSMWGPYNVY
MAV_3310|M.avium_104 ------------
MAB_2743|M.abscessus_ATCC_1997 ------------
Mb1824|M.bovis_AF2122/97 ------------
Rv1796|M.tuberculosis_H37Rv ------------
MSMEG_1600|M.smegmatis_MC2_155 ------------
Mvan_1509|M.vanbaalenii_PYR-1 ------------
MMAR_0950|M.marinum_M ------------
MUL_0701|M.ulcerans_Agy99 ------------
Mflv_0760|M.gilvum_PYR-GCK ------------
TH_2316|M.thermoresistible__bu LMQTDLSSLVEV