For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTPTQTLAQPRFARGALLAFAGLTAFSVAACGTHGDTNAASTRSSAPTTSSSAPHGEHNEKGHVSGLIVS VSGNSVEVTKQNGNATVSFGQSTKVSEIFAAQLSDVTAGSCIAAMTPPDSNPPTARRVMMWPSADGSCVR EHPKEQQSTSPAPTSGKRPEHEGIRGTVASVSGNAITVTGTDPNGTATGPTTVTVNNDTTYAKRAPVDQN AIATGKCMAAHGTDDASGNLQATSITLRAARDGSCDGGGHEGHHQR
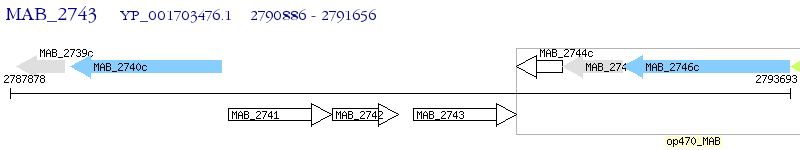
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2743 | - | - | 100% (256) | hypothetical protein MAB_2743 |
| M. abscessus ATCC 19977 | MAB_2741 | - | 6e-35 | 37.50% (248) | hypothetical protein MAB_2741 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3310 | - | 3e-50 | 45.78% (249) | 27 kDa lipoprotein antigen |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2743|M.abscessus_ATCC_1997 MTPTQTLAQPRFARGALLAFAGLTAFSVAACGTHGDTNAASTRSSAPTTS
MAV_3310|M.avium_104 ----MSASCHGLARVAVFAVAGATAVSLSACGTSNKSSPTSTSTSTSTVT
: : :** *::*.** **.*::**** ..:..:** :*:.*.:
MAB_2743|M.abscessus_ATCC_1997 SSAPHGEHNEKGHVSGLIVSVSGNSVEVTKQ-NGNATVSFGQSTKVSEIF
MAV_3310|M.avium_104 STSTSAAPNAEAKVSGLIASVAGNSIQVTKEDNGTAAVNFTSTTKITEAV
*::. . * :.:*****.**:***::***: **.*:*.* .:**::* .
MAB_2743|M.abscessus_ATCC_1997 AAQLSDVTAGSCIAAMTPPDSNP---PTARRVMMWPSADGSCVREH-PKE
MAV_3310|M.avium_104 PAGLPDVTPGSCVSVKPTEGSAPGQPVTAASVKISQSVNGACPKPHQPTP
.* *.***.***::. .. .* * ** * : *.:*:* : * *.
MAB_2743|M.abscessus_ATCC_1997 QQSTSPAP---TSGKRPEHEGIRGTVASVSGNAITVTGTDPNGTATGPTT
MAV_3310|M.avium_104 GGSSSPAPSLPPSQAPAKPALVRGSVASVSGNTINVTGTDPSGNTT-QTT
*:**** .* .: :**:*******:*.******.*.:* **
MAB_2743|M.abscessus_ATCC_1997 VTVNNDTTYAKRAPVDQNAIATGKCMAAHGTDDASGNLQATSITLRAARD
MAV_3310|M.avium_104 VTVDDKTKYTKQSSANTDAITPGKCLSARGTNDSGGALQATSIQLRQAVD
***::.*.*:*::..: :**:.***::*:**:*:.* ****** ** * *
MAB_2743|M.abscessus_ATCC_1997 GSCDGGGHEGHHQR
MAV_3310|M.avium_104 GKCGKPKQPGHGG-
*.*. : **