For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKHLTLATISVALSLALVSCGHDHKSGDKTSSSTSTATSSKSSTAPAAQAKYTIADYVKDNHISETPVHH GDPGPNVDLPVPAGWQLNQNSGTSYGGIVQSQPADPSDPPTVSALFSKLTGDVDPAKIIQYAPGEVQNLP GYSGSGDGSASTLGGYQAWQLGGTYQRGGKTRVIAQKTVVIPSQGAVYVLQLNADGLESDQGPLMDATHI IDEQTTITG
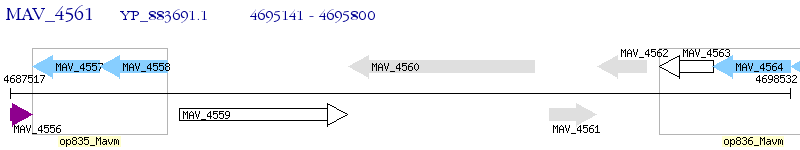
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4561 | - | - | 100% (219) | LpqN protein |
| M. avium 104 | MAV_1154 | - | 2e-08 | 25.76% (198) | LpqT protein |
| M. avium 104 | MAV_3310 | - | 6e-06 | 29.87% (154) | 27 kDa lipoprotein antigen |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0598c | lpqN | 2e-79 | 62.56% (227) | lipoprotein LpqN |
| M. gilvum PYR-GCK | Mflv_1917 | - | 4e-47 | 44.44% (225) | hypothetical protein Mflv_1917 |
| M. tuberculosis H37Rv | Rv0583c | lpqN | 2e-79 | 62.56% (227) | lipoprotein LpqN |
| M. leprae Br4923 | MLBr_00246 | lpqT | 1e-09 | 25.98% (204) | putative lipoprotein |
| M. abscessus ATCC 19977 | MAB_1145c | - | 9e-12 | 25.73% (206) | putative lipoprotein LpqT precursor |
| M. marinum M | MMAR_0950 | lpqN | 9e-79 | 63.00% (227) | lipoprotein, LpqN |
| M. smegmatis MC2 155 | MSMEG_5456 | - | 2e-57 | 52.02% (223) | MK35 lipoprotein |
| M. thermoresistible (build 8) | TH_0934 | lpqN | 1e-48 | 45.00% (220) | PROBABLE CONSERVED LIPOPROTEIN LPQN |
| M. ulcerans Agy99 | MUL_0701 | lpqN | 2e-78 | 62.67% (225) | lipoprotein, LpqN |
| M. vanbaalenii PYR-1 | Mvan_4816 | - | 9e-49 | 46.33% (218) | LpqN |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0598c|M.bovis_AF2122/97 ----MKHFTAAVATVALSLALAGCSFNIKT----DSAPTTSPTTTSPTTS
Rv0583c|M.tuberculosis_H37Rv ----MKHFTAAVATVALSLALAGCSFNIKT----DSAPTTSPTTTSPTTS
MMAR_0950|M.marinum_M ----MKDLTAVLATAALSLTLVACGSGTTS----DNKTSTSTSTSTSTST
MUL_0701|M.ulcerans_Agy99 ----MKDLTAVLATAALSLTLVACGSGTTS----DNK--TSTSTSTSTST
MAV_4561|M.avium_104 ----MKHLTLATISVALSLALVSCGHDHKS----GDK-------TSSSTS
Mflv_1917|M.gilvum_PYR-GCK MIIANRAGAAAVA-VAVGIGLSACGSQTTG-DATTAQETPSASETSSTSS
Mvan_4816|M.vanbaalenii_PYR-1 MTIATRAGVAAIAAVALGIGLSACGSDST-------DEKPSAASSSATS-
MSMEG_5456|M.smegmatis_MC2_155 MITTFRSGGAVLAACALATALSACGSDVTAGQATTGDQTISATSTAAQTS
TH_0934|M.thermoresistible__bu -MSRTARRATRIAAVAAAAALAACGSTADD---QTVMPPDTGGATASATM
MLBr_00246|M.leprae_Br4923 MQAIRLGLHTAAAVVTLSISAVSCGTKTPD-----------------YQL
MAB_1145c|M.abscessus_ATCC_199 -----MRSAALAAAAVAAFAMIGCGSHNPG-----------------QPT
. . .*.
Mb0598c|M.bovis_AF2122/97 TTTTSATTSAQAAGPNYTIADYIRDNHIQETPVHHGDPGSPTIDLPVPDD
Rv0583c|M.tuberculosis_H37Rv TTTTSATTSAQAAGPNYTIADYIRDNHIQETPVHHGDPGSPTIDLPVPDD
MMAR_0950|M.marinum_M STTTSATSPTPGAQANYTIADYIRDNDIQETPVHHGDPGSPNIDLPVPDG
MUL_0701|M.ulcerans_Agy99 STTTSATSPTPGAQANYTIADYIRDNDIQETPVHHGDPGSPNIDLPVPDG
MAV_4561|M.avium_104 TATSSKSSTAPAAQAKYTIADYVKDNHISETPVHHGDPG-PNVDLPVPAG
Mflv_1917|M.gilvum_PYR-GCK TTPAAPTSPSPAAAPGITIQQYVDENGITSTPVAKGEPGAPTIQLPIPPG
Mvan_4816|M.vanbaalenii_PYR-1 ETAASPASPTPAAGPGITLAQYIEENGIAETPVLRGDPGAPTINLPFPQG
MSMEG_5456|M.smegmatis_MC2_155 AKAAPEPIAEGTSGPHYTIVDYIRDNGITEMPVHRGDPGTPVLDIPIPPG
TH_0934|M.thermoresistible__bu TTTSAAQAGAQAGASGYTIVDYIRDHDIDETTARPGDPGAPNVSLPTPPG
MLBr_00246|M.leprae_Br4923 ILSKSSTTTTTTPDKPIPLPQYLESIGVTGQQVAPSSLPGLTVSIPTPPG
MAB_1145c|M.abscessus_ATCC_199 GSASSTPESAAPPAKPASMNEYLHSVGVTVTPVSPESPANVRVDVPLPQG
. . .: :*: . : . . :.:* * .
Mb0598c|M.bovis_AF2122/97 WRLLP-ESSRAPYGGIVYTQPADPNDPPTIVAILSKLTGDIDPAKVL-QF
Rv0583c|M.tuberculosis_H37Rv WRLLP-ESSRAPYGGIVYTQPADPNDPPTIVAILSKLTGDIDPAKVL-QF
MMAR_0950|M.marinum_M WQLLP-EGSGAPYGGIAYTQPADPSDPPTIVAILSKLTGNVEAAKIL-EF
MUL_0701|M.ulcerans_Agy99 WQLLP-EGSGAPYGGIAYTQPADPSDPPTIVAILSKLTGNVEAAKIL-EF
MAV_4561|M.avium_104 WQLN--QNSGTSYGGIVQSQPADPSDPPTVSALFSKLTGDVDPAKII-QY
Mflv_1917|M.gilvum_PYR-GCK WEPAGPQTPEGAYDALVLAG--PSPNKPTIVAFVDKLTGPVDTAKIL-EY
Mvan_4816|M.vanbaalenii_PYR-1 WEAAGPRAPQGAYDAIIYTAGPPGAQPATIVAFVTKLTGNVDSAKII-EY
MSMEG_5456|M.smegmatis_MC2_155 WADAGADTPEWAWSAMVSTDPAFADDPPMIIALMSRLTGDVDPAKIL-EY
TH_0934|M.thermoresistible__bu WSNATDRAPGWAWAALVSGDPAVADDPPSIVVLMSKLTGDVDADRVL-EF
MLBr_00246|M.leprae_Br4923 WS-----PYSNPNITPETLIIAKSGKYPTARLVAFKLRGDFDPTQVI-KH
MAB_1145c|M.abscessus_ATCC_199 WENLG--ALDPAYLIADKPSGADQGRTPSAVVYLIKLGGPLDARKVISEH
* . . :* * .:. ::: :.
Mb0598c|M.bovis_AF2122/97 APGELKNLPGFQG-SGDGSAATLGGFSAWQLGGSYSKNGKLRTVAQKTVV
Rv0583c|M.tuberculosis_H37Rv APGELKNLPGFQG-SGDGSAATLGGFSAWQLGGSYSKNGKLRTVAQKTVV
MMAR_0950|M.marinum_M APGELKNLPGYEG-SNNGSASTLSGFQAWQLGGSYTKNGKQRAVAQKTVV
MUL_0701|M.ulcerans_Agy99 APGELKNLPGYEG-SNNGSASTLSGFQAWQLGGSYTKNGKQRAVAQKTVV
MAV_4561|M.avium_104 APGEVQNLPGYSG-SGDGSASTLGGYQAWQLGGTYQRGGKTRVIAQKTVV
Mflv_1917|M.gilvum_PYR-GCK APNETIALPGFEG-ATEGKPSTLSGFDATQIGGFYTRDGVTRMLAQKTTV
Mvan_4816|M.vanbaalenii_PYR-1 APNETRNLPGFEG-ADKGEPSNLGGFDATQIGGFYTKDGVNHLVAQKTVV
MSMEG_5456|M.smegmatis_MC2_155 APNEIKNLPGYDGEGSEGTADELSGFDAYQIGGMYVRDGQQRLIAQKTVV
TH_0934|M.thermoresistible__bu APAELRNLPGFDG--GAGTATTLSGFDAVQIGGRYDKDGVERVVGQKTVV
MLBr_00246|M.leprae_Br4923 GNDDAQLFENFRQ--LDVSTANYNGFPSAMIQGSYDLEGRRLHAWNRIVI
MAB_1145c|M.abscessus_ATCC_199 GFADAQNTQNFQK--IASSLDDYQGYPSAAIEGTYENQGVRVHAWNRDVI
. : .: *: : : * * * :: .:
Mb0598c|M.bovis_AF2122/97 IPSQG---AVFVLQLNADALDDETMTLMDAANVIDEQTTITP-
Rv0583c|M.tuberculosis_H37Rv IPSQG---AVFVLQLNADALDDETMTLMDAANVIDEQTTITP-
MMAR_0950|M.marinum_M IPSQD---GLFVLQLDADALDSEQDVLMDATGVIDDQTKITP-
MUL_0701|M.ulcerans_Agy99 IPSQD---GLFVLQLDADALDSEQDVLMDATGVIDDQTKITP-
MAV_4561|M.avium_104 IPSQG---AVYVLQLNADGLESDQGPLMDATHIIDEQTTITG-
Mflv_1917|M.gilvum_PYR-GCK IPTDG---AVFVLKVTAEGTDDFAMPLMDATAAIDEQTTITP-
Mvan_4816|M.vanbaalenii_PYR-1 IPAQD---GLFVLKLIADGTEEVAYPLMEATSAIDEQTTITP-
MSMEG_5456|M.smegmatis_MC2_155 IPGQD---GLYVLQLNADGTEDQLGTLLDATSVIDEKTVITP-
TH_0934|M.thermoresistible__bu IPAAD---AVYVMQLNADAPAEHAEQLLNATRFLDEQTRITF-
MLBr_00246|M.leprae_Br4923 PTGPPPSKQQYLVQLTITSLANEAVAQSNDIEAIIRGFVVAPK
MAB_1145c|M.abscessus_ATCC_199 VPDGP---LHYLVQFTVTTTEAQSAALSQDVAALTGGLKITKR
. ::::. : : ::