For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SADPNVSDPTTIAARGPARRRVPGPTLAQVFDTRINALNVWRLVLASGVILQHSWPLTGRKVHPPYDQLF PQVWVDGFFAISGFLIAASWVRNPRLREYVAARALRILPGLWVCLVVIAFVIAPIGVAIQGGSAAKLLMS RAPIEYVLHNAVLHVFFAGIDGTPRHVPFPGVWDGVLWTLIFEIFCYIAIAVAGVAGLLKRRWPAVVVFI LLVVFSALVAYPVEVQTVFQMIGRFALVFAAGTLLHQFQDKIPARWSLVALSAVIVLAAGLLMPNYRVLA AIPLAYAVVVSGALIHHERLRLRTDLSYGVYIYAWPMQQLLAICGLSFLPPVVFAVVAVVVTLPLAALSW FLVEKRALSLKSRIKRRARGWGAVVSSGSRISTPGS*
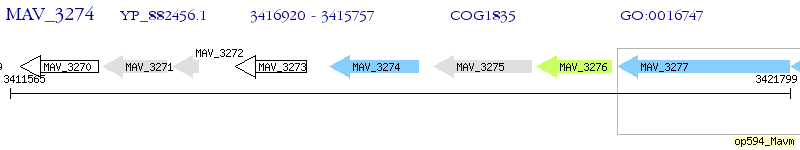
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3274 | - | - | 100% (387) | putative acyltransferase |
| M. avium 104 | MAV_4410 | - | e-109 | 56.62% (355) | AtfA_1 |
| M. avium 104 | MAV_4409 | - | e-103 | 55.75% (348) | putative acyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0233 | - | 7e-05 | 24.63% (341) | integral membrane acyltransferase |
| M. gilvum PYR-GCK | Mflv_4779 | - | 2e-05 | 25.79% (349) | acyltransferase 3 |
| M. tuberculosis H37Rv | Rv0228 | - | 7e-05 | 24.63% (341) | integral membrane acyltransferase |
| M. leprae Br4923 | MLBr_01101 | - | 2e-05 | 24.08% (191) | putative acyltransferase |
| M. abscessus ATCC 19977 | MAB_4106c | - | 1e-124 | 60.83% (360) | acetyltransferase |
| M. marinum M | MMAR_4188 | - | 3e-08 | 26.34% (205) | integral membrane acyltransferase |
| M. smegmatis MC2 155 | MSMEG_0390 | - | 1e-127 | 64.14% (343) | putative acyltransferase |
| M. thermoresistible (build 8) | TH_4330 | - | 2e-06 | 25.15% (326) | PUTATIVE acyltransferase 3 |
| M. ulcerans Agy99 | MUL_4492 | - | 2e-08 | 26.83% (205) | integral membrane acyltransferase |
| M. vanbaalenii PYR-1 | Mvan_0251 | - | 3e-07 | 24.41% (340) | acyltransferase 3 |
CLUSTAL 2.0.9 multiple sequence alignment
TH_4330|M.thermoresistible__bu --------------MPGTLVAMAVTAEQVAGNPGDSSGASPRDLSHEVSG
Mvan_0251|M.vanbaalenii_PYR-1 ---------------------MTVTAA-----PADPS---------VVGG
Mb0233|M.bovis_AF2122/97 -------------MGPADESGAPIRPQTPHRHTVLVTNG------QVVGG
Rv0228|M.tuberculosis_H37Rv -------------MGPADESGAPIRPQTPHRHTVLVTNG------QVVGG
MMAR_4188|M.marinum_M ---------------------MTLPEA------TDAQGG------LEQIS
MUL_4492|M.ulcerans_Agy99 ---------------------MTLPEA------TDAQGG------LEQIS
MLBr_01101|M.leprae_Br4923 ---------------------MTLSEE------RDAQSG------LKQVS
MAB_4106c|M.abscessus_ATCC_199 -------------MTKAGATGQAKG-------------------LTLGQV
MSMEG_0390|M.smegmatis_MC2_155 ----------------------------------------------MGKV
MAV_3274|M.avium_104 MSADPNVSDPTTIAARGPARRRVPG-------------------PTLAQV
Mflv_4779|M.gilvum_PYR-GCK -------------MVTRVDQWREASAR--------------------RRR
TH_4330|M.thermoresistible__bu TRSFLPAVEGLRACAALGVIVTHVAFQTG-HTSGAVGRLLGRFDLAVAVF
Mvan_0251|M.vanbaalenii_PYR-1 TRGFLPAVEGLRACAAIGVVVTHVAFQTG-QSGGVVGRVLGRFDLAVAVF
Mb0233|M.bovis_AF2122/97 TRGFLPAVEGMRACAAVGVVVTHVAFQTG-HSSGVGGRLFGRFDLAVAVF
Rv0228|M.tuberculosis_H37Rv TRGFLPAVEGMRACAAVGVVVTHVAFQTG-HSSGVGGRLFGRFDLAVAVF
MMAR_4188|M.marinum_M HVDRVASLTGIRAVAALLVVATHAAYTTGKYTHGYWGLVSARMEIGVPIF
MUL_4492|M.ulcerans_Agy99 HVDRVASLTGIRAVAALLVVTTHAAYTTGKYTHGYWGLVSARMEIGVPIF
MLBr_01101|M.leprae_Br4923 HVDRVASLTGIRAVAALLIVGTHAAYTTGKYTHGYWGLVGARLEIGVPIF
MAB_4106c|M.abscessus_ATCC_199 FDPRNNALNAWRLALAVSVIFWHSWPLTG-RTITYRPFEQLIEQVGVDGF
MSMEG_0390|M.smegmatis_MC2_155 FDPRNNALNALRLLLAVGVILWHSWPLSG-HGVTYKPLEQLLEQVWVDGF
MAV_3274|M.avium_104 FDTRINALNVWRLVLASGVILQHSWPLTG-R-KVHPPYDQLFPQVWVDGF
Mflv_4779|M.gilvum_PYR-GCK RAERANHRLDVQGLRAVAVLGVFAHFLTGWPRGGFVG---------IDIF
: * :: . :* : *
TH_4330|M.thermoresistible__bu FALSGFLLWRGHAAAVRGLRPTPATGRYLRSRVVRIMPGYLVAVVVILLL
Mvan_0251|M.vanbaalenii_PYR-1 FALSGFLLWRGHAAAARGLRSRPTTGHYLRSRVVRIMPGYLVAVVVIMTL
Mb0233|M.bovis_AF2122/97 FAVSGFLLWRGHAAAARDLRSHPRTGPYLRSRVARIMPAYVVAVVVILSL
Rv0228|M.tuberculosis_H37Rv FAVSGFLLWRGHAAAARDLRSHPRTGPYLRSRVARIMPAYVVAVVVILSL
MMAR_4188|M.marinum_M FVLSGFLLFRPWVHSAVTGAPPPSLRRYTAHRVRRIMPAYVVVVLIAYVI
MUL_4492|M.ulcerans_Agy99 FVLSGFLLFRPWVHSAVTGAPPPSLRRYTAHRVRRIMPAYVVVVLIAYVI
MLBr_01101|M.leprae_Br4923 FALSGFLLFSPWVKSAATGSPPPSVSRYAWHRVRRIMPAYVITVLFAYAL
MAB_4106c|M.abscessus_ATCC_199 FAVSGFLITASWLR-------KPKLRDFAIARGLRIFPGLWVCLAIIAFV
MSMEG_0390|M.smegmatis_MC2_155 FAISGFLITSSWLR-------NPRLRDYATARVLRIFPGLWVCLLIIAFV
MAV_3274|M.avium_104 FAISGFLIAASWVR-------NPRLREYVAARALRILPGLWVCLVVIAFV
Mflv_4779|M.gilvum_PYR-GCK FVIAGFFVTESLLRTAADTG-SPSLRTFYVGRVRRILPAATAVLILTVAA
*.::**:: * : * **:*. : .
TH_4330|M.thermoresistible__bu LPGAT------ADLTVWLANLTLTQIYVP---------LTLTSGLTQMWS
Mvan_0251|M.vanbaalenii_PYR-1 LPDVN------ADFTVWLANLTLTQIYVP---------LTLTSGLTQMWS
Mb0233|M.bovis_AF2122/97 LPDADH-----ASLTVWLANLTLTQIYVP---------LTLTGGLTQMWS
Rv0228|M.tuberculosis_H37Rv LPDADH-----ASLTVWLANLTLTQIYVP---------LTLTGGLTQMWS
MMAR_4188|M.marinum_M YHYRIAGPNPGHSWLGLVRNLTLTQIYTDGY-----LGSYLHQGLTQMWS
MUL_4492|M.ulcerans_Agy99 YHYRIAGPNPGHSWMGLVRNLTLTQIYTDGY-----LGSYLHQGLTQMWS
MLBr_01101|M.leprae_Br4923 YHFRAAGPNPGHSWMGLVRNLTLTQIYTDGY-----LGSYLHQGLTQMWS
MAB_4106c|M.abscessus_ATCC_199 IAPTS----------VLIQGGSVSDLFAS---------------HKPIEY
MSMEG_0390|M.smegmatis_MC2_155 MAPIG----------VLLQGGSVSGLMSS---------------YAPIEY
MAV_3274|M.avium_104 IAPIG----------VAIQGGSAAKLLMS---------------RAPIEY
Mflv_4779|M.gilvum_PYR-GCK TALVMPDDLRDVGVDALFAFFFVANWHFAASGVDAASAAESASPLLHYWP
. :
TH_4330|M.thermoresistible__bu LSVEMTFYLVLPLLALLARQVPVRARIP-------VLLATAVLSLFWVRI
Mvan_0251|M.vanbaalenii_PYR-1 LSVEMSFYLALPLLALLARRLPVRARIP-------AIVVVAAASLFWVRI
Mb0233|M.bovis_AF2122/97 LSVEVAFYAALPVLALLGRRIPVGARVP-------AIAALAALSWAWGWL
Rv0228|M.tuberculosis_H37Rv LSVEVAFYAALPVLALLGRRIPVGARVP-------AIAALAALSWAWGWL
MMAR_4188|M.marinum_M LAVEASFYVLLPFLAYLLLVVISRRRWQPK-LVVGALLAMATITPGWLVL
MUL_4492|M.ulcerans_Agy99 LAVEASFYVLLPFLAYLLLVVISRRRWQPK-LVVGALLAMATITPGWLVL
MLBr_01101|M.leprae_Br4923 LAVEVAFYVALPLLAYLLLVLLCQRRWQPR-LVLVVLAGMLLISPGWLIL
MAB_4106c|M.abscessus_ATCC_199 VINNGLLNVYYAGVDGTPRGVPWPGVWNG------SIWTLVFEMICYIAV
MSMEG_0390|M.smegmatis_MC2_155 VLNNAVLNVYYAGINGTPQGIPWPGVWNG------SLWTLVFEMICYIAV
MAV_3274|M.avium_104 VLHNAVLHVFFAGIDGTPRHVPFPGVWDG------VLWTLIFEIFCYIAI
Mflv_4779|M.gilvum_PYR-GCK LAVEEQFFIVWPVLILGITAIAVRKAWTDERWRAHLAVAVGVVVIASLAW
: : : . : :
TH_4330|M.thermoresistible__bu PFGPDSGLN-PWNWPPAFFSWFAAGMILAELTVTRVGWFHRLARRRVLMG
Mvan_0251|M.vanbaalenii_PYR-1 PFDPDSGHN-LWNWPPAFFSWFAAGMVLAELTVGPVGWAHRLARRRVLMA
Mb0233|M.bovis_AF2122/97 PLDAGSGIN-PLTWPPAFFSWFAAGMLLAEWAYSPVGLPHRWARRRVAMA
Rv0228|M.tuberculosis_H37Rv PLDAGSGIN-PLTWPPAFFSWFAAGMLLAEWAYSPVGLPHRWARRRVAMA
MMAR_4188|M.marinum_M IHTVSLFPDGARLWLPSYLAWFLGGMLLTVLQAMKVQCYALAALP-----
MUL_4492|M.ulcerans_Agy99 IHTVSLFPDGARLWLPSYLAWFLGGMLLTVLQAMKVQCYALAALP-----
MLBr_01101|M.leprae_Br4923 VHPDHWFPDGARLWLPTYLAWFLGGMILAVLQVMGVRCYAFGAIP-----
MAB_4106c|M.abscessus_ATCC_199 AVLGVAGLLKRRWVVPAAFVFFLAATAYLSYPVFAATSIPQMVAR-----
MSMEG_0390|M.smegmatis_MC2_155 AVLGIVGLLKRRWVIPAIFVLTLCATAYFSYPVFAMQTIPQMVAR-----
MAV_3274|M.avium_104 AVAGVAGLLKRRWPAVVVFILLVVFSALVAYPV-EVQTVFQMIGR-----
Mflv_4779|M.gilvum_PYR-GCK AAVQTASSPSWAYFDTFARVWELGVGALLALAVGTLSRIPDPIRP-----
TH_4330|M.thermoresistible__bu MFALAAFLVAASPLVGLEGLAPGSIVQVTLKTVMGALLAAALLAPLVLDR
Mvan_0251|M.vanbaalenii_PYR-1 VLAVTAFLIAASPLAGREGLHPGSVGQVTLKTAMGAVVAGALLAPLVLDR
Mb0233|M.bovis_AF2122/97 VTALLGYLVAASPLAGPEGLVPGTAAQFAVKTAMGSLVAFALVAPLVLDR
Rv0228|M.tuberculosis_H37Rv VTALLGYLVAASPLAGPEGLVPGTAAQFAVKTAMGSLVAFALVAPLVLDR
MMAR_4188|M.marinum_M -LATICYFIVATPIAGAPTTSPSALTEAVVKACFYTVIATLAVAPLALGD
MUL_4492|M.ulcerans_Agy99 -LATICYFIVATPIAGAPTTSPSALTEAVVKACFYTVIATLAVAPLALGD
MLBr_01101|M.leprae_Br4923 -LAIICYFIVSTPIAGAPTTSPASLGEALIKVGFYAAIATLAVAPLTLGD
MAB_4106c|M.abscessus_ATCC_199 ---FAVMFAAGALLNQYKDILPARWSLVALSVAIVLVSGLLE-NYRDVAA
MSMEG_0390|M.smegmatis_MC2_155 ---FAVMFAAGALIHQYRDVIPARWSLVGIAVVLVLASGFLS-NYRVIGA
MAV_3274|M.avium_104 ---FALVFAAGTLLHQFQDKIPARWSLVALSAVIVLAAGLLMPNYRVLAA
Mflv_4779|M.gilvum_PYR-GCK -----ILSWTGLTLIGAAFVLVDGGSGFPVPWALLPVAGAALVIAAGVGR
.. : : : . :
TH_4330|M.thermoresistible__bu PDTPHRFLASAPMVTLGRWSYG---IFVWHLAALAMVFP-MIGEFPFNGN
Mvan_0251|M.vanbaalenii_PYR-1 IDTPHRILGSPVMVTLGRWSYG---LFVWHLAALAMVFP-IIGAFPFNGH
Mb0233|M.bovis_AF2122/97 PDTSHRLLGSPAMVTLGRWSYG---LFIWHLAALAMVFP-VIGAFPFTGR
Rv0228|M.tuberculosis_H37Rv PDTSHRLLGSPAMVTLGRWSYG---LFIWHLAALAMVFP-VIGAFPFTGR
MMAR_4188|M.marinum_M QGWYSQLLASRPMVWLGEISYE---IFLVHLVTMEFAMVYVVRYRVYTGS
MUL_4492|M.ulcerans_Agy99 QGWYSQLLASRPMVWLGEISYE---IFLVHLVTMEFAMVYVVRYWVYTGS
MLBr_01101|M.leprae_Br4923 LGWYSWLLSSRPMVWLGEISYE---IFLIHLVTMEFAMVYVVQDHVYTGS
MAB_4106c|M.abscessus_ATCC_199 LPLAYAVIVSGSLIHKPRLNLRNDLSYGTYIYAYPIQQLLVVSGLASL-N
MSMEG_0390|M.smegmatis_MC2_155 LPLAYAVIVAGALLRNRRLNLRNDLSYGVYIYAFPVQQLLAILGFANS-Q
MAV_3274|M.avium_104 IPLAYAVVVSGALIHHERLRLRTDLSYGVYIYAWPMQQLLAICGLSFL-P
Mflv_4779|M.gilvum_PYR-GCK EPELQGFLRNRVSTYLGDISFALYLVHWPVIVLLAAVMDRSVFYYASVVT
.: . : :
TH_4330|M.thermoresistible__bu MPVVLVLTVLFTIAIAAVSYGLVEQPCRAAYRRWEMRRARPDAPAPPLDS
Mvan_0251|M.vanbaalenii_PYR-1 FTVVLVLTVLFGFAIAAVSYALVESPCRNALRRWERR----NHPTP-LDS
Mb0233|M.bovis_AF2122/97 MPTVLVLTLIFGFAIAAVSYALVESPCREALRRWERR----NEPIS-VGE
Rv0228|M.tuberculosis_H37Rv MPTVLVLTLIFGFAIAAVSYALVESPCREALRRWERR----NEPIS-VGE
MMAR_4188|M.marinum_M MLNLFIATMVITIPLAWVLHRFTRVRSE----------------------
MUL_4492|M.ulcerans_Agy99 MLNLFIATMVITIPLAWVLHRFTRVRSE----------------------
MLBr_01101|M.leprae_Br4923 MLNLIVATLVLTIPLAWLLHRCTRMRS-----------------------
MAB_4106c|M.abscessus_ATCC_199 LLLFFVIATVATVPLAAFSWFVVEKRAIALKSRLTRKQA--VTVAERPPI
MSMEG_0390|M.smegmatis_MC2_155 PFLFFVIATAATVPLAALSWFLVEKRAIALKSRLKRRRT--DTVADTTQK
MAV_3274|M.avium_104 PVVFAVVAVVVTLPLAALSWFLVEKRALSLKSRIKRRARGWGAVVSSGSR
Mflv_4779|M.gilvum_PYR-GCK LAFGLALAVHHFVENPARDATVQGLRQAREDRRHGLHHTELSTKIAGVAA
: . .
TH_4330|M.thermoresistible__bu AVTSVATPISR--------------------
Mvan_0251|M.vanbaalenii_PYR-1 SVSDEIEPARAR-------------------
Mb0233|M.bovis_AF2122/97 LQADAIAP-----------------------
Rv0228|M.tuberculosis_H37Rv LQADAIAP-----------------------
MMAR_4188|M.marinum_M -------------------------------
MUL_4492|M.ulcerans_Agy99 -------------------------------
MLBr_01101|M.leprae_Br4923 -------------------------------
MAB_4106c|M.abscessus_ATCC_199 PLDRDAA------------------------
MSMEG_0390|M.smegmatis_MC2_155 VSDRGAG------------------------
MAV_3274|M.avium_104 ISTPGS-------------------------
Mflv_4779|M.gilvum_PYR-GCK LILITFSVISYAMSPEAIDDPAPIAPTAPSP