For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVPPNLVDLAEAAGLSAIAYGPDMHDFWSDEFIRDFFKNFLRGPVTLIREAWEPVLKNWQEMSSALMSVG AGADLLFTGQLYQDLAINVADYYGIPMATLHYIPMRPNGQLLPRVPGPLVRTGMAVYDWMCWRMNKKAED RQRRELGLPTATVASPRRIAERGSLEIQAYDEVCFPGLAEEWAKWGGQRPFVGSLTMELPTDFDDEVLSW IGQGTPPICFGFGSMPVAESPADTVEMIGAACAELGERALICSGWSDFSNVPHPDHVKVVGVVNYTKIFP ACRAVVHHGGSGTTAAGLRAGIPSLILWTAGDQPLWGSHIKRLKVGTSRRFSATTPETLVADLRKILAPE YLTRARELSSRMSKPAESAGRAVDLLEEFARSGRCIPRAEAQRSR*
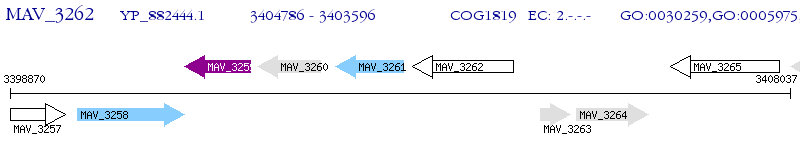
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3262 | - | - | 100% (396) | glycosyl transferase |
| M. avium 104 | MAV_3265 | - | e-147 | 65.04% (389) | glycosyl transferase |
| M. avium 104 | MAV_3258 | - | e-145 | 63.87% (382) | glycosyltransferase family protein 28 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1551 | - | 1e-133 | 60.57% (388) | glycosyltransferase |
| M. gilvum PYR-GCK | Mflv_3984 | - | 6e-07 | 24.02% (333) | UDP-glucuronosyl/UDP-glucosyltransferase |
| M. tuberculosis H37Rv | Rv1524 | - | 1e-133 | 60.57% (388) | glycosyltransferase |
| M. leprae Br4923 | MLBr_02348 | - | 1e-121 | 56.51% (384) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_4107c | - | 1e-132 | 60.61% (391) | glycosyltransferase GtfA |
| M. marinum M | MMAR_2353 | - | 1e-121 | 55.41% (379) | UDP-glycosyltransferase |
| M. smegmatis MC2 155 | MSMEG_0389 | - | 1e-133 | 60.56% (393) | glycosyl transferase |
| M. thermoresistible (build 8) | TH_1989 | - | 9e-53 | 65.25% (141) | Glycosyltransferase family protein 28 |
| M. ulcerans Agy99 | MUL_1529 | - | 1e-109 | 53.54% (353) | UDP-glycosyltransferase |
| M. vanbaalenii PYR-1 | Mvan_4034 | - | 3e-05 | 26.47% (136) | glycosyl transferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1551|M.bovis_AF2122/97 -MKFVVASYGTRGDIEPCAAVGLELQRRGHDVCLAVPPNLIGFVETAGLS
Rv1524|M.tuberculosis_H37Rv -MKFVVASYGTRGDIEPCAAVGLELQRRGHDVCLAVPPNLIGFVETAGLS
MMAR_2353|M.marinum_M -MKFALAINGTRGDVEPCAALGRELSRRGHEVRVAVPPNLTGLAAAAGLE
MUL_1529|M.ulcerans_Agy99 -----MSNRALHSGGNYRAEATKDAS--------QYPPNLTGVAAAAGLE
MLBr_02348|M.leprae_Br4923 -MKFTLAASGSRGDVEPFAALGLELQRRGHEVRIGVPPDMLRFVESAGLA
MAV_3262|M.avium_104 ---------------------------------MAVPPNLVDLAEAAGLS
MAB_4107c|M.abscessus_ATCC_199 -MKFVLASYGTRGDIEPSVVVARELQRRGHDVVMAVPPDLISFTESAGLE
MSMEG_0389|M.smegmatis_MC2_155 -MKFVLASYGTRGDIEPSVVVGRELQSRGHTVLMAVPPDLIEFTEAANLR
TH_1989|M.thermoresistible__bu --------------------------------------------------
Mflv_3984|M.gilvum_PYR-GCK -MRVAVVAGPDPGHAFPALELCRRFRAAGDAPTLLTGTRWLDVARGEGVD
Mvan_4034|M.vanbaalenii_PYR-1 MPEILIATCPPVGHLSPLLNVARGLVARGDRVTVLTSARHADKIRAVGAE
Mb1551|M.bovis_AF2122/97 AVAYGSRDSQEQLDEQ------FLHNAWK--LQNPIKLLREAMAPVTEGW
Rv1524|M.tuberculosis_H37Rv AVAYGSRDSQEQLDEQ------FLHNAWK--LQNPIKLLREAMAPVTEGW
MMAR_2353|M.marinum_M AVPYGP-DTDELLNDE-----DNLQEFWK--PQTSIKLVREGISDLRQAW
MUL_1529|M.ulcerans_Agy99 AVPYCP-DTDELLNDE-----DNLQEFWK--PQTSIKLVREGISDLRQAW
MLBr_02348|M.leprae_Br4923 AVAYGP-DTQEFLARDT---YSQWRQWWK--ILPPIKALQQ----LRQAW
MAV_3262|M.avium_104 AIAYGP-DMHDFWSDE------FIRDFFKNFLRGPVTLIREAWEPVLKNW
MAB_4107c|M.abscessus_ATCC_199 TVSYGL-DTKTWLDVYRNFWTFFFHTFWK--VREIRTMWRQMWELSDECW
MSMEG_0389|M.smegmatis_MC2_155 TVAYGL-DTKTWLDIYRNFWTFALHRFWK--AREIRSLWREMWDLSDECW
TH_1989|M.thermoresistible__bu --------------------------------------------------
Mflv_3984|M.gilvum_PYR-GCK AVELDGLDPTAFDDDT----------------DAGEKIHRRAARMAVLNR
Mvan_4034|M.vanbaalenii_PYR-1 PRPLPHAADYDDSTLDADLPGRAATAGLARINFDIESIFVRPLPHQFSAL
Mb1551|M.bovis_AF2122/97 AELSAMLTPVAAGADLLLTG-QIYQE----VVANVAEHHGIPLAALHFYP
Rv1524|M.tuberculosis_H37Rv AELSAMLTPVAAGADLLLTG-QIYQE----VVANVAEHHGIPLAALHFYP
MMAR_2353|M.marinum_M SDMAGTLKTLADGADLVLTG-MIHQG----IALNAAEYYGIPLATMHFLP
MUL_1529|M.ulcerans_Agy99 SDMAGTLKTLADGADLVLTG-MIHQG----IALNAAEYYGIPLATMHFLP
MLBr_02348|M.leprae_Br4923 ADMATDLKSLADGADLVMTG-VVYQG----VVANVAEYYGIPFGVLHFVP
MAV_3262|M.avium_104 QEMSSALMSVGAGADLLFTG-QLYQD----LAINVADYYGIPMATLHYIP
MAB_4107c|M.abscessus_ATCC_199 AQMNTTLMSAAQGADVLFAG-QSYQE----PAANVAEYHDIPLVTLHHIP
MSMEG_0389|M.smegmatis_MC2_155 AQMNATLRSVAADADVLFAG-QSYQE----PAANVAEFYDVPLVTLHHIP
TH_1989|M.thermoresistible__bu --------------------------------------------------
Mflv_3984|M.gilvum_PYR-GCK ARIGELSPDLVVSDSITVCGGLAADL----LGVPWVELNTHPLYRPSRGL
Mvan_4034|M.vanbaalenii_PYR-1 QELLVTDRFDAVIADALFLGTLPLLLGDPAKRPPILSYSTTPLFLTSRDT
Mb1551|M.bovis_AF2122/97 VRANGEIAFPARLPAPLVRSTIT-AIDWLYWRMTKGVEDAQRRELGLPKA
Rv1524|M.tuberculosis_H37Rv VRANGEIAFPARLPAPLVRSTIT-AIDWLYWRMTKGVEDAQRRELGLPKA
MMAR_2353|M.marinum_M SRVNSRVIP--ILSPRLNRSVIS-ALWWVHWRVTKNPEEAQRRELGMAKA
MUL_1529|M.ulcerans_Agy99 SRVNSRVIP--ILSPRLNRSVIS-ALWWVHWRVTKNPEEAQRRELGMAKA
MLBr_02348|M.leprae_Br4923 ARVNGKIIP--SLPSPLNRAILA-TVWRAHWLLAKKPEDAQRRELGLPKA
MAV_3262|M.avium_104 MRPNGQLLP--RVPGPLVRTGMA-VYDWMCWRMNKKAEDRQRRELGLPTA
MAB_4107c|M.abscessus_ATCC_199 MRPNGQLVT--ILPSRLGRAAMR-AFDWVSWRLNKKVEDAQRRELRLPKA
MSMEG_0389|M.smegmatis_MC2_155 MRPNGRLVT--FLPPVLARASMT-AFDWFAWRLNKKVEDAQRRDLGLPKA
TH_1989|M.thermoresistible__bu --------------------------------------------------
Mflv_3984|M.gilvum_PYR-GCK PPVGSGLAPGVGLRGRARDAVLR-ALTARSWRAGLRQRAAARVEIGLPGD
Mvan_4034|M.vanbaalenii_PYR-1 APAGPGIAPMPGMIGRLRNRVLARATQSVLLRPGQRAADRMLEAMGLPEL
Mb1551|M.bovis_AF2122/97 STPAPRRMAVRGSLEIQAYDALCFPGLAAEW---GGRRPFVGALT-MESA
Rv1524|M.tuberculosis_H37Rv STPAPRRMAVRGSLEIQAYDALCFPGLAAEW---GGRRPFVGALT-MESA
MMAR_2353|M.marinum_M SSSSTSRILKGRSLEIQAYDELCFPGLAAEWAEWGDRRPFVGALT-LEMP
MUL_1529|M.ulcerans_Agy99 SSSSTSRILKGRSLEIQAYDELCFPGLAAEWAEWGDRRPFVGALT-LEMP
MLBr_02348|M.leprae_Br4923 TSLSTRRIVKRGALEIQAYDELCFPGLAAEWAEYGDRRPFVGALT-LELP
MAV_3262|M.avium_104 TVASPRRIAERGSLEIQAYDEVCFPGLAEEWAKWGGQRPFVGSLT-MELP
MAB_4107c|M.abscessus_ATCC_199 SSPSPQRIAERRSLEIQGYDSVCFPGLATEWAKWNGLRPFVGSLT-MAMT
MSMEG_0389|M.smegmatis_MC2_155 TGPSTQRIADRMSLEVQAYDEVCFPGLAEEWAKWGELRPYVGTLT-MELT
TH_1989|M.thermoresistible__bu --------------------------------------------------
Mflv_3984|M.gilvum_PYR-GCK DPGPVRRLIAT------------LPALEVPRPDWPAEAVVVGPLH-FEPT
Mvan_4034|M.vanbaalenii_PYR-1 PVFILDSAVLADRVIVP-----TVPEFEYRRSDLPSHVRFVGPVSPLPGS
Mb1551|M.bovis_AF2122/97 TDADDEVASWIAADTPPIYFGFGSMP-IGS-LADRVAMISAACAELGERA
Rv1524|M.tuberculosis_H37Rv TDADDEVASWIAADTPPIYFGFGSMP-IGS-LADRVAMISAACAELGERA
MMAR_2353|M.marinum_M TESDNEVLSWIASGTPPIYFGFGSMP-VES-PAATIAMISAACAELGERA
MUL_1529|M.ulcerans_Agy99 TESDNEVLSWIASGTPPIYFGFGSMP-VES-PAATIAIISAACAELGERA
MLBr_02348|M.leprae_Br4923 TAADNEVLSWIAAGTPPIYFGFGSMP-IVS-PTDTVAMIAAACADLGERA
MAV_3262|M.avium_104 TDFDDEVLSWIGQGTPPICFGFGSMP-VAESPADTVEMIGAACAELGERA
MAB_4107c|M.abscessus_ATCC_199 QEADEEVASWIAGGTPPIFFGFGSMP-VES-PSAALEMIGSACAELGERA
MSMEG_0389|M.smegmatis_MC2_155 TPADEEVSSWICGGTPPICFGFGSMP-VES-PADTVAMISSVCAELGERA
TH_1989|M.thermoresistible__bu -------------------------------------MFASVCSRLGERA
Mflv_3984|M.gilvum_PYR-GCK T----ELLAVPAGGGPLVVVAPSTAT-TGAGGMAELALNTLVPGDTLPPG
Mvan_4034|M.vanbaalenii_PYR-1 DFVAPPWWGELNAGRPVVHVTQGTIDNADLTRLIEPTIEALADEDVTVVA
: . .
Mb1551|M.bovis_AF2122/97 LICSGPSDATGIPQFDHVKVVRVVSHAAVFPTCRAVVHHGGAGTTAAGLR
Rv1524|M.tuberculosis_H37Rv LICSGPSDATGIPQFDHVKVVRVVSHAAVFPTCRAVVHHGGAGTTAAGLR
MMAR_2353|M.marinum_M LICTGSNDLGGLQAPENVKLVSGVSHAAVFPACRAVVHHGGAGTTAAGMR
MUL_1529|M.ulcerans_Agy99 LICTGSNDLGGLQAPENVKLVSGVSHAAVFPACCAVVHHGGAGTTAAGMR
MLBr_02348|M.leprae_Br4923 LISVKPKDLTQVPKFDHVKIVTSVSHAAVFPACRAVVHHGGAGTTAASLR
MAV_3262|M.avium_104 LICSGWSDFSNVPHPDHVKVVGVVNYTKIFPACRAVVHHGGSGTTAAGLR
MAB_4107c|M.abscessus_ATCC_199 LIGAGWSDFGGAQLPEHVKVVGAVNYATVFPACRAVVHHGGSGTTAASLR
MSMEG_0389|M.smegmatis_MC2_155 LICAGWSDFSDVALADHVKVVGAVNYATVFPGCRAVVHHGGSGTTAASLR
TH_1989|M.thermoresistible__bu LICAGGTDIDGVPEYGHVKVVGAVNYARVFPACRAIVHHGGSGTTAAVLR
Mflv_3984|M.gilvum_PYR-GCK ARVAVSQLDGPDSAVPPWAVVGLGRQDELLARADLLICGGGHGTVAKSLL
Mvan_4034|M.vanbaalenii_PYR-1 TTGGRPISQIRIPLPANTFVAKYVPHDVLLPMVDVMVTNGGYGAVQRALS
:. ::. :: ** *:. :
Mb1551|M.bovis_AF2122/97 AGIPTLILWVTSDQPIWAAQIKQLKVGRGRRFSSATKESLIADLRTILA-
Rv1524|M.tuberculosis_H37Rv AGIPTLILWVTSDQPIWAAQIKQLKVGRGRRFSSATKESLIADLRTILA-
MMAR_2353|M.marinum_M AGIPTLILWIWIEQPIWLAQIKRLKIGSGRRLASTTKETLVSDLRCILG-
MUL_1529|M.ulcerans_Agy99 AGIPTLILWIWIEQPIWLAQIKRLKIGSGRRLASTTKETLVSDLRCILG-
MLBr_02348|M.leprae_Br4923 AGVPTLILWIFIEQPVWAAQIKRLKVGAGRRFSATTQRSLAADLRTILA-
MAV_3262|M.avium_104 AGIPSLILWTAGDQPLWGSHIKRLKVGTSRRFSATTPETLVADLRKILA-
MAB_4107c|M.abscessus_ATCC_199 AGVPTLILSMDANQTLWGGQVKKLKVGTTRRFSTTTRESLVADLRRILA-
MSMEG_0389|M.smegmatis_MC2_155 AGLPTLVLSMDANQALWGAQIKRLKVGTTRRFSKTNRRTLIADLRTILD-
TH_1989|M.thermoresistible__bu AGVPSLVTWSSADQPYWGNQVKRLKVGTSRRFSHTTPDTLVADLRRILT-
Mflv_3984|M.gilvum_PYR-GCK AGVPMVVVPGGGDQ--WEIANRLVRQGSAQLVRPLSAESLAAAVREVLGS
Mvan_4034|M.vanbaalenii_PYR-1 DGVPVVVAGHTEDKPEVAARVRHFGVGIDLRTGTPTPTQVRRAVRKVLHE
*:* :: :: : . * . : :* :*
Mb1551|M.bovis_AF2122/97 PDYVTRAREIASRMTKPAASVTATADLLEDAARRAR-----------
Rv1524|M.tuberculosis_H37Rv PDYVTRAREIASRMTKPAASVTATADLLEDAARRAR-----------
MMAR_2353|M.marinum_M PDYVARARTIANEMSKPGESATRAADLLENEVSVS------------
MUL_1529|M.ulcerans_Agy99 PDYVARP---------PG--LRRC-----------------------
MLBr_02348|M.leprae_Br4923 PQYATRAREVANRMSKPDESVNAAADLLEDKASRNKSRTRYQDC---
MAV_3262|M.avium_104 PEYLTRARELSSRMSKPAESAGRAVDLLEEFARSGRCIPRAEAQRSR
MAB_4107c|M.abscessus_ATCC_199 SDYADRARELAAGMTKPGESAVAAADAMERFAQARCSA---------
MSMEG_0389|M.smegmatis_MC2_155 PAYAARAQALARGMTRPAESVVRAADVIEAFARSRCSA---------
TH_1989|M.thermoresistible__bu PDYAVRARAVAARMTDPADSVVRAADLFETAVECAPGVAHRE-----
Mflv_3984|M.gilvum_PYR-GCK PSYREAAVRAGAGVADVADPVRVCHDALGAPRKLGPCG---------
Mvan_4034|M.vanbaalenii_PYR-1 PGFRTKAGWLRSAYAAYDSVAEIAKLVDEAVAQRTQPLQSA------
. : .