For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LGFDRLPEHIVDKRVTELMDLSGKKAVVTGAGGDGLGQAIANRLGGLGADIALIGRTLEKVQRRGREVEE RWGVKTVAISADMSDWDQVHNAVREAHWQLGGLDIMVNNPVMVAGGLFETQTKEQIDFTVLGSLTMMMYG AHAALQFLLPQGSGKIINIGSVGGRIQQRGLVVYNACKAGVIGFTRNLAHEVALRGVNVLGVAPGIMLNP QLKQYVLDPQDDQERAGRDAMIEAITQQVQLGRASLPEEAANMVAFLATEAADYLCGQTIDVAGGQWMG*
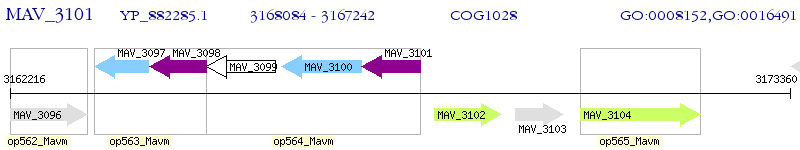
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3101 | - | - | 100% (280) | oxidoreductase, short chain dehydrogenase/reductase family protein |
| M. avium 104 | MAV_1572 | - | 5e-30 | 32.20% (264) | short chain dehydrogenase |
| M. avium 104 | MAV_5088 | fabG | 2e-26 | 29.84% (258) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1385 | fabG | 5e-30 | 32.09% (268) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1832 | - | 1e-125 | 78.49% (279) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1350 | fabG | 5e-30 | 32.09% (268) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-20 | 29.02% (255) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4178c | - | 3e-26 | 30.19% (265) | short chain dehydrogenase |
| M. marinum M | MMAR_4033 | fabG | 1e-30 | 33.08% (266) | 3-oxoacyl- |
| M. smegmatis MC2 155 | MSMEG_2206 | fabG | 2e-28 | 32.45% (265) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. thermoresistible (build 8) | TH_3693 | - | 1e-27 | 31.82% (264) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3898 | fabG | 3e-29 | 32.69% (260) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_1975 | fabG | 9e-29 | 32.09% (268) | 3-ketoacyl-(acyl-carrier-protein) reductase |
CLUSTAL 2.0.9 multiple sequence alignment
TH_3693|M.thermoresistible__bu -----------------------------VSLLAGQ--------TAVVTG
Mvan_1975|M.vanbaalenii_PYR-1 -----------------------MRMGEQVSLLTGQ--------TAVITG
MSMEG_2206|M.smegmatis_MC2_155 -------------------------------MLTGQ--------TAVVTG
Mb1385|M.bovis_AF2122/97 ----------------------------MASLLNAR--------TAVITG
Rv1350|M.tuberculosis_H37Rv ----------------------------MASLLNAR--------TAVITG
MMAR_4033|M.marinum_M ----------------------------MSVSLLNG-------QIAVVTG
MUL_3898|M.ulcerans_Agy99 ----------------------------MCGAHRDRG-----YVYPVVTG
MLBr_01807|M.leprae_Br4923 ----------------------MTDAAAKVSAAPGRGKPAFVSRSILVTG
MAV_3101|M.avium_104 -----------MLGFDRLPEHIVDKRVTELMDLSGKK-------AVVTGA
Mflv_1832|M.gilvum_PYR-GCK MTTHNDAEAIAALNLDRLPHHIVGKRVTDLMDLSGKK-------AFVTGA
MAB_4178c|M.abscessus_ATCC_199 ---------------------------MSLLNFDGKV-------VVVSGI
:
TH_3693|M.thermoresistible__bu GARGLGYAIAERFIAEGARVVIGDIDESATRAAADSLGGAD--VAVGMRC
Mvan_1975|M.vanbaalenii_PYR-1 GAQGLGFAIAQRFVEEGARVVLGDVNLDATVAAAEKLGGED--VARAVRC
MSMEG_2206|M.smegmatis_MC2_155 GAQGLGLAIAKRFISEGARVVLGDLNSEATEAAVEELGGSE--VAAAVRC
Mb1385|M.bovis_AF2122/97 GAQGLGLAIGQRFVAEGARVVLGDVNLEATEVAAKRLGGDD--VALAVRC
Rv1350|M.tuberculosis_H37Rv GAQGLGLAIGQRFVAEGARVVLGDVNLEATEVAAKRLGGDD--VALAVRC
MMAR_4033|M.marinum_M GAQGLGLAIAERFVSEGARVVLGDVNLEATEAAAKQLGGGE--VAVAVRC
MUL_3898|M.ulcerans_Agy99 GAQGLGLAIAERFVSEGARVVLGDVNLEATEAAAKQLGGGE--VAVAVRC
MLBr_01807|M.leprae_Br4923 GNRGIGLAVARRLVADGHRVAVTHR---ASVVPDWFFG---------VEC
MAV_3101|M.avium_104 GGDGLGQAIANRLGGLGADIALIGRTLEKVQRRGREVEERWGVKTVAISA
Mflv_1832|M.gilvum_PYR-GCK GGDGLGHAIANRLAGSGADIALVGRTREPLERRAAEIAQRWGVNTYPVQA
MAB_4178c|M.abscessus_ATCC_199 G-PGLGAALAIKFAEAGADVVLAARTQSRLDEVARQIAATG-RRALAVAT
* *:* *:. :: * :.: . :
TH_3693|M.thermoresistible__bu DVTRSDEVEALVGAAVKRFGSLDVMVNNAGITRDATMRKMTEQQFDEVIA
Mvan_1975|M.vanbaalenii_PYR-1 DVTDSAEVDALIAAAVDGFGSLDIMVNNAGITRDATMRKMTEEQFDQVIA
MSMEG_2206|M.smegmatis_MC2_155 DVTSSADVDALVQAAVERFGGLDIMVNNAGITRDATLRKMTEEQFDQVIA
Mb1385|M.bovis_AF2122/97 DVTQADDVDILIRTAVERFGGLDVMVNNAGITRDATMRTMTEEQFDQVIA
Rv1350|M.tuberculosis_H37Rv DVTQADDVDILIRTAVERFGGLDVMVNNAGITRDATMRTMTEEQFDQVIA
MMAR_4033|M.marinum_M DVTKADEVETLLQTALERFGGLDIMVNNAGITRDATMRKMTEEQFDQVID
MUL_3898|M.ulcerans_Agy99 DVTKADEVETLLQAALERFGGLDIMVNNAGITRDATMRKMTEEQFDQVID
MLBr_01807|M.leprae_Br4923 DVTDNDAIGAAFKAVEEHQGPVEVLVANAGITSDMLIMRMSEEQFQKVID
MAV_3101|M.avium_104 DMSDWDQVHNAVREAHWQLGGLDIMVNNPVMVAGGLFETQTKEQIDFTVL
Mflv_1832|M.gilvum_PYR-GCK DMSKWDQIHRGVAEAHDKLGRLDIMINNPVMVAAGPFEKQTKEQIDLTVL
MAB_4178c|M.abscessus_ATCC_199 DITDEASAANLVERATEEFGGIDVLVNNAFSIPSMKSLEKTDYQHIRDSV
*:: . . * ::::: *. :. *
TH_3693|M.thermoresistible__bu VHLKGTWNGLRAAAAIMREQKRGAIVNMSSISGKVGMVGQTNYSAAKAGI
Mvan_1975|M.vanbaalenii_PYR-1 VHLKGTWNGLRAAAAVMREQKRGAIVNMSSISGKVGMVGQTNYSAAKAGI
MSMEG_2206|M.smegmatis_MC2_155 VHLKGTWNGTKAAAAIMRENKRGAIVNMSSISGKVGLIGQTNYSAAKAGI
Mb1385|M.bovis_AF2122/97 VHLKGTWNGTRLAAAIMRERKRGAIVNMSSVSGKVGMVGQTNYSAAKAGI
Rv1350|M.tuberculosis_H37Rv VHLKGTWNGTRLAAAIMRERKRGAIVNMSSVSGKVGMVGQTNYSAAKAGI
MMAR_4033|M.marinum_M VHLKGTWNGIRLAAAIMRENKRGAIVNMSSVSGKVGMVGQTNYSAAKAGI
MUL_3898|M.ulcerans_Agy99 VHLKGTWNGIRLAAAIMRENKRGAIVNMSSVSGKVGMVGQTNYSAAKAGI
MLBr_01807|M.leprae_Br4923 VNLTGVFRVAKLASRNMIKQRFGRYIFMGSVVGLSGVPGQANYAASKSGL
MAV_3101|M.avium_104 GSLTMMMYGAHAALQFLLPQGSGKIINIGSVGGRIQQRGLVVYNACKAGV
Mflv_1832|M.gilvum_PYR-GCK GSLTMMMYGAHAALEYLLPQGSGVIINIGSVGGRVQQRGLTVYNACKSGV
MAB_4178c|M.abscessus_ATCC_199 ELTVVGTLRLTQRVAAALAQSKGAVVNINSMVIRHSQVRYGSYKIAKAAL
. * : :.*: * .*:.:
TH_3693|M.thermoresistible__bu VGMTKAASKELAYLGVRVNAIQPGLIR------------SAMTEAMPQRI
Mvan_1975|M.vanbaalenii_PYR-1 VGMTKAASKELAYLGVRVNAIQPGLIR------------SAMTEAMPQRI
MSMEG_2206|M.smegmatis_MC2_155 VGMTKAAAKELAYLGVRVNAIQPGLIR------------SAMTEAMPQRI
Mb1385|M.bovis_AF2122/97 VGMTKAAAKELAHLGIRVNAIAPGLIR------------SAMTEAMPQRI
Rv1350|M.tuberculosis_H37Rv VGMTKAAAKELAHLGIRVNAIAPGLIR------------SAMTEAMPQRI
MMAR_4033|M.marinum_M VGMTKAAAKELAHVGVRVNAIAPGLIR------------SAMTEAMPQRI
MUL_3898|M.ulcerans_Agy99 VGMTKAAAKELAHVGVRVNAIAPGLIR------------SAMTEAMPQRI
MLBr_01807|M.leprae_Br4923 IGMARSLARELGSRSITANVVAPGFID------------TEMTRAMDSKY
MAV_3101|M.avium_104 IGFTRNLAHEVALRGVNVLGVAPGIMLNPQLKQYVLDPQDDQERAGRDAM
Mflv_1832|M.gilvum_PYR-GCK IGFTRNLAHEVAPRGVNVFCVAPGIMIKEQMKQYLFDPQDDQQRAGRGAI
MAB_4178c|M.abscessus_ATCC_199 LAMSQSLATELGPQGVRVNSVVPGYMWGENLKGY-FEHQAKKYGGTVEQI
:.::: : *:. .: . : ** : .
TH_3693|M.thermoresistible__bu WDEKLAEIPMGRAGEPEEVAKVALFLASDLSSYMTGTVLEVTGGRHL--
Mvan_1975|M.vanbaalenii_PYR-1 WDSKVAEVPMGRAGEPEEVANVALFLASDLSSYMTGTVLEVTGGRHL--
MSMEG_2206|M.smegmatis_MC2_155 WDEKLAEIPMGRAGEPDEVAKVALFLASDLSSYMTGTVLEVTGGRHV--
Mb1385|M.bovis_AF2122/97 WDQKLAEVPMGRAGEPSEVASVAVFLASDLSSYMTGTVLDVTGGRFI--
Rv1350|M.tuberculosis_H37Rv WDQKLAEVPMGRAGEPSEVASVAVFLASDLSSYMTGTVLDVTGGRFI--
MMAR_4033|M.marinum_M WDQKLAEVPMGRAGEPSEVASVALFLACDLSSYMTGTVLDVTGGRFI--
MUL_3898|M.ulcerans_Agy99 WDQKLAEVPMGRAGEPSEVASVALFLACDLSSYMTGTVLDVTGGRFI--
MLBr_01807|M.leprae_Br4923 QEAALQAIPLGRIGAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH
MAV_3101|M.avium_104 IEAITQQVQLGRASLPEEAANMVAFLATEAADYLCGQTIDVAGGQWMG-
Mflv_1832|M.gilvum_PYR-GCK IESITHQVQLGRASLPEEAANMVAFLASEAASYMCGQTIDVAGGQWMG-
MAB_4178c|M.abscessus_ATCC_199 YEYTAANSDLKRLPTTDEVANAVLFLASDLASGITGQALDVNCGEFHA-
: : * .* * . *** : :. : * .: * *