For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNRKRVVVAGLGDVGVLAAIRLSRHADVVGISAKPALVSGQELGVRLSRPHDWARDYWIPFDRFRRLDGV RTVQATLTGVDLAARTVFGRGDDGAPVAEPYDALVICTGVSNGFWRRPTLQSAAEIGADLRDAHERLAAA GSVIVVGGGAAAVSSALNMARTWPDKRIDLYFPGETALSGHHPRAWRRLHARLTRLGVGVHPGHRAVLPD GFAGDELTSRPVQWSTGQPPAAADAVLWAIGRVRPNTGWLPAELLDERGFVRVTPQLRVPGHPGVFAVGD VAATDPLRSSARNRGDALVARNVVAEFTGRPLRGFRAPRRRWGSLVGIQPDGLEVFLPSGHTVRFPSWSV QRVVMPLIVRWGMYGGVRRNDPLGRETATPPTL
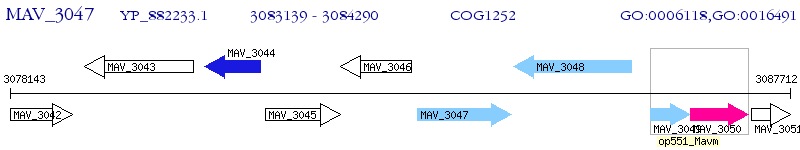
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3047 | - | - | 100% (383) | pyridine nucleotide-disulphide oxidoreductase, putative |
| M. avium 104 | MAV_2867 | - | 2e-08 | 25.71% (350) | DoxD family protein/pyridine nucleotide-disulfide |
| M. avium 104 | MAV_1130 | - | 3e-07 | 25.85% (325) | dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1842c | - | 2e-09 | 27.09% (358) | dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1066 | - | 1e-150 | 70.14% (365) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. tuberculosis H37Rv | Rv1812c | - | 2e-09 | 27.09% (358) | dehydrogenase |
| M. leprae Br4923 | MLBr_02061 | - | 8e-06 | 24.56% (281) | putative dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2429c | - | 4e-09 | 23.78% (349) | NADH dehydrogenase (NDH) |
| M. marinum M | MMAR_2587 | ndh1 | 1e-178 | 79.89% (373) | NADH dehydrogenase Ndh1 |
| M. smegmatis MC2 155 | MSMEG_6582 | - | 1e-138 | 65.95% (370) | pyridine nucleotide-disulphide oxidoreductase domain-containing |
| M. thermoresistible (build 8) | TH_4200 | - | 2e-08 | 24.30% (321) | PUTATIVE dihydrolipoamide dehydrogenase |
| M. ulcerans Agy99 | MUL_3173 | ndh1 | 1e-177 | 79.62% (373) | NADH dehydrogenase Ndh1 |
| M. vanbaalenii PYR-1 | Mvan_5760 | - | 1e-154 | 71.08% (370) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2587|M.marinum_M ---------------MN-----------RKRVVVAG---LG----DVGVL
MUL_3173|M.ulcerans_Agy99 ---------------MN-----------RKRVVVAG---LG----DVVVL
MAV_3047|M.avium_104 ---------------MN-----------RKRVVVAG---LG----DVGVL
Mflv_1066|M.gilvum_PYR-GCK ---------------MTQ----------GRRVVIAG---LG----DVGVL
Mvan_5760|M.vanbaalenii_PYR-1 ---------------MS-----------GRRVVIAG---MG----DVGVL
MSMEG_6582|M.smegmatis_MC2_155 ---------------MTRTTPVGSAVVTPKRVVIAG---LG----DAGVL
Mb1842c|M.bovis_AF2122/97 ----------------------------MTRVVVIG---SG----FAGLW
Rv1812c|M.tuberculosis_H37Rv ----------------------------MTRVVVIG---SG----FAGLW
MLBr_02061|M.leprae_Br4923 ---------------MNAQPAATAAQDRRHQVVIIG---SG----FGGLN
MAB_2429c|M.abscessus_ATCC_199 MSTTPDAAGAAAPVAANPQPKIQDALTPKHRVVIIG---SG----FGGLT
TH_4200|M.thermoresistible__bu VEAVSHYDVVVLGAGPGGYVAAIRAAQLGQKTAIIEPRYWGGVCLNVGCI
:..: *
MMAR_2587|M.marinum_M IAIRLSRHCDVVGISAKPALVSGQE----LGVRLTRPHDWARDYWIAFDK
MUL_3173|M.ulcerans_Agy99 IAIRLSRHCDVVGISAKPALVSGQE----LGVRLTRPHDWARDYWIAFDK
MAV_3047|M.avium_104 AAIRLSRHADVVGISAKPALVSGQE----LGVRLSRPHDWARDYWIPFDR
Mflv_1066|M.gilvum_PYR-GCK TAIKLARHADVVGISAKPGLVSGQE----LGWRLARPGDWARHNWIPFSR
Mvan_5760|M.vanbaalenii_PYR-1 TAIKLADHADVIGISAKPGLVSGQE----LGWRLARPDDWARHNWIPFGR
MSMEG_6582|M.smegmatis_MC2_155 TAIRLARRHDVVGVSLNPGLVSGQE----LGVRLARPDDWARDYWISFDR
Mb1842c|M.bovis_AF2122/97 AALGAARRLDELAVPAGTVDVMVVSNKPFHDIRVRNYEADLSACRIPLGD
Rv1812c|M.tuberculosis_H37Rv AALGAARRLDELAVLAGTVDVMVVSNKPFHDIRVRNYEADLSACRIPLGD
MLBr_02061|M.leprae_Br4923 AAKKLKRANVDIKLIARTTHHLFQPLLYQVATGIISEGEIAPPTRVVLRK
MAB_2429c|M.abscessus_ATCC_199 AAKTLKRANADVTLIARTTHHLFQPLLYQVATGIISEGEIAPPTRQILKD
TH_4200|M.thermoresistible__bu PSKALLRNAELASIFTRDAKLFGIQGEVTFDYGAAFDRSRKVADGRVSGV
: :
MMAR_2587|M.marinum_M FRRLDRVHTIQAELTGVDLAARSVFVRCDDGTTFAEAYDALVISTGVSNG
MUL_3173|M.ulcerans_Agy99 FRRLDRVHTIQAELTGVDLAARSVFVRCDDGTTFAEAYDALVISTGVSNG
MAV_3047|M.avium_104 FRRLDGVRTVQATLTGVDLAARTVFGRGDDGAPVAEPYDALVICTGVSNG
Mflv_1066|M.gilvum_PYR-GCK FRGLDRVRTVHGTLTAVDLDARTVAVSCADGESTVVPYDVLVIATGVSNG
Mvan_5760|M.vanbaalenii_PYR-1 FRGLDKVRTVHGTLTGVDLTARTVAVTLPDGSSTEVVYDALVVATGVANG
MSMEG_6582|M.smegmatis_MC2_155 LRALDRVRTVHGALTGADLDTRTITVTDADGTEHHEPYDALVIATGVTNG
Mb1842c|M.bovis_AF2122/97 VLGPAGVAHVTAEVTAIDADGRRVTTSTG----ASYSYDRLVLASGSHVV
Rv1812c|M.tuberculosis_H37Rv VLGPAGVAHVTAEVTAIDADGRRVTTSTG----ASYSYDRLVLASGSHVV
MLBr_02061|M.leprae_Br4923 QR---NIQVLLGNVTHIDLANQCVVSELLG-HTYETLYDSLIVAAGAGQS
MAB_2429c|M.abscessus_ATCC_199 QD---NAQVVLGDVTSIDLEKQVVHSSLLG-HDYSTPYDSLIVAAGAGQS
TH_4200|M.thermoresistible__bu HFLMKKNKITEIHGYGRFTDAHTIEVELNEGGTEQVTFDNAIIATGSST-
: : :* ::.:*
MMAR_2587|M.marinum_M FWRRP----------TLQSPADIGAELQAAHHRLAAAA---------SVI
MUL_3173|M.ulcerans_Agy99 FWRRP----------TLQSPADIGAELQAAHHRLAAAA---------SVI
MAV_3047|M.avium_104 FWRRP----------TLQSAAEIGADLRDAHERLAAAG---------SVI
Mflv_1066|M.gilvum_PYR-GCK FWRHP----------GLQTSEQVDADLRNPHERLSAAQ---------SVA
Mvan_5760|M.vanbaalenii_PYR-1 FWRQP----------TLQTAAQIGADLHGPHEKLAAAR---------SVI
MSMEG_6582|M.smegmatis_MC2_155 FWRRP----------VLQSPDDVHQGLREAHDRLSAAT---------SVA
Mb1842c|M.bovis_AF2122/97 KPALPGLAEFGFDVDTYDGAVRLQQHLQGLAGGPLTS-------AAATVV
Rv1812c|M.tuberculosis_H37Rv KPALPGLAEFGFDVDTYDGAVRLQQHLQGLAGGPLTS-------AAATVV
MLBr_02061|M.leprae_Br4923 YFGNDQFAEFAPGMKSIDDALELRGRILSAFEQAERSNDPERREKLLTFT
MAB_2429c|M.abscessus_ATCC_199 YFGNDHFAEWAPGMKSIDDALELRGRILGAFEQAERSSDPVRRRKLMTFT
TH_4200|M.thermoresistible__bu ---------------KLVPGTSLSENVVTYEKLIMTRELPG------SIV
: : :.
MMAR_2587|M.marinum_M VIGGGAAAVSSAVNMATTWPDKRIDLYFPGE-----RPLTAYHPRAWQRI
MUL_3173|M.ulcerans_Agy99 AIGGGAAAVSSAVNMATTWPDKRIDLYFPGE-----RPLTAYHPRAWQRI
MAV_3047|M.avium_104 VVGGGAAAVSSALNMARTWPDKRIDLYFPGE-----TALSGHHPRAWRRL
Mflv_1066|M.gilvum_PYR-GCK IVGGGAAAVSSAAQIATTWPDKRVELYFPGD-----RALAGHHPRTWKTV
Mvan_5760|M.vanbaalenii_PYR-1 VVGGGAAAVSSAAQIATTWPDKRVELYFPGE-----RALAGHHPRTWTTV
MSMEG_6582|M.smegmatis_MC2_155 VIGGGAAAVSSAANLAARWPDKHVGLYHPGN-----EILPQHHRRTRNRI
Mb1842c|M.bovis_AF2122/97 VVGAGLTGIETACELPG----RLHALFARGDGVTPRVVLIDHNPFVGSDM
Rv1812c|M.tuberculosis_H37Rv VVGAGLTGIETACELPG----RLHALFARGDGVTPRVVLIDHNPFVGSDM
MLBr_02061|M.leprae_Br4923 VVGAGPTGVEMAGQIAELADHTLKGAFRRIDSTKARVVLLDAAPAVLPPM
MAB_2429c|M.abscessus_ATCC_199 VVGAGPTGVEMAGQIAELANETLKGTFRHIDPTDARVILLDAAPAVLPPF
TH_4200|M.thermoresistible__bu IAGAGAIGMEFGYIMANYGVDVTVVEFLP-------RVLPNEDPEVSREI
*.* .:. . :. : * . .
MMAR_2587|M.marinum_M RGRLTGLGVGLHPGHRAVTPDAFTGDEITNAPVRWTTEQ---PAAHADAV
MUL_3173|M.ulcerans_Agy99 RGRLTGLGVGLHPGHRAVTPDAFTGDEITNAPVRWTTGQ---PAAHADAV
MAV_3047|M.avium_104 HARLTRLGVGVHPGHRAVLPDGFAGDELTSRPVQWSTGQ---PPAAADAV
Mflv_1066|M.gilvum_PYR-GCK RRRLTEAGVVLRPGHRAQLVPGFTGAELTTDPVQWTTGQ---DPATADVV
Mvan_5760|M.vanbaalenii_PYR-1 RRRLHEAGVVLRPGHRADLEPGFIGDELTTGPVRWSTGQ---PPSTADAV
MSMEG_6582|M.smegmatis_MC2_155 TQRLNALGVDVHPGHRAIVPDGFACDEITDDPVHFSTGQ---SPAKAEAV
Mb1842c|M.bovis_AF2122/97 GLSARPVIEQALLDNGVETRTGVSVAAVSPGGVTLSSGE----RLAAATV
Rv1812c|M.tuberculosis_H37Rv GLSARPVIEQALLDNGVETRTGVSVAAVSPGGVTLSSGE----RLAAATV
MLBr_02061|M.leprae_Br4923 GEKLGQRAAARLQKMGVEIQLSAMVTDVDRNGITVQDSDGTVRRIESACK
MAB_2429c|M.abscessus_ATCC_199 GPKLGDKARKRLEKLGVEIQLSAMVTDVDRNGLTVKHADGSIERIESWCK
TH_4200|M.thermoresistible__bu EKQFKKLGVKIHTGTKVESITDNGAGGSVVVKVSKDGES---SELKADKV
. : . :
MMAR_2587|M.marinum_M LWAIGKVRPNTGWLPE----QLLDDHGFVRVTPQLRVPGRRDVFAVGDVA
MUL_3173|M.ulcerans_Agy99 LWAIGKVRPNTGWLPE----QLLDDHGFVRVTPQLRVPGRRDVFAVGDVA
MAV_3047|M.avium_104 LWAIGRVRPNTGWLPA----ELLDERGFVRVTPQLRVPGHPGVFAVGDVA
Mflv_1066|M.gilvum_PYR-GCK LWAIGRVRPNTGWLPG----ELLDADGFVRVTPQLQVPGRPEVFAVGDVA
Mvan_5760|M.vanbaalenii_PYR-1 LWAIGRVRPNTGWLPT----ELLDADGFVRVTPTLQVPGHPEVFAVGDVA
MSMEG_6582|M.smegmatis_MC2_155 LWAIGRVTPNTGWLPR----AILDDDGFVRVTPRLQVPGVPGVFAVGDVA
Mb1842c|M.bovis_AF2122/97 VWCAGMRASRLTEQLP----VARDRLGRLQVDDYLRVIGVPAMFAAGDVA
Rv1812c|M.tuberculosis_H37Rv VWCAGMRASRLTEQLP----VARDRLGRLQVDDYLRVIGVPAMFAAGDVA
MLBr_02061|M.leprae_Br4923 VWSAGVSASRLGRDLAEQSLVELDWAGRVKVLPDLSIPGYRNVFVVGDMA
MAB_2429c|M.abscessus_ATCC_199 VWSAGVSASPLGKNLAEQSGVELDRAGRVKVGPDLSIPGHPNVFVVGDMM
TH_4200|M.thermoresistible__bu LQAIGFAPNVDGYGLDRAG-VRLDDRQAIAIDDYMRTS-VPHIYAIGDVT
: . * * : : : ::. **:
MMAR_2587|M.marinum_M ASDP------------------------------------LRSSARNRGD
MUL_3173|M.ulcerans_Agy99 ASDP------------------------------------LRSSARNRGD
MAV_3047|M.avium_104 ATDP------------------------------------LRSSARNRGD
Mflv_1066|M.gilvum_PYR-GCK ATDP------------------------------------LRSSARNRAD
Mvan_5760|M.vanbaalenii_PYR-1 ATDP------------------------------------LRSSARNRAD
MSMEG_6582|M.smegmatis_MC2_155 ATDA------------------------------------LRSSARNRAD
Mb1842c|M.bovis_AF2122/97 AAR-----------------------------MDDEHLSVMSCQHGRPMG
Rv1812c|M.tuberculosis_H37Rv AAR-----------------------------MDDEHLSVMSCQHGRPMG
MLBr_02061|M.leprae_Br4923 AIDGVPGVAQGAIQGAKYVANNIKAELGEANPAEREPFQYFDKGSMATVS
MAB_2429c|M.abscessus_ATCC_199 AVDGVPGVAQGAIQGGRYAAKQIKAELKGRSPAERAPFKFFDKGSMATVS
TH_4200|M.thermoresistible__bu AKLQLAHVAEAMG-----VVAAETIAGVETLPLGDYRMMPRATFCQPQVA
*
MMAR_2587|M.marinum_M ALVANNIRAEFTGKPLRSYRAPRRRWGSLLGI-QPDGLEVFLPTGHSLRL
MUL_3173|M.ulcerans_Agy99 ALVANNIRAEFTGKPLRSYRAPRRRWGSLLGI-QPDGLEVFLPTGHSLRL
MAV_3047|M.avium_104 ALVARNVVAEFTGRPLRGFRAPRRRWGSLVGI-QPDGLEVFLPSGHTVRF
Mflv_1066|M.gilvum_PYR-GCK GLLAGNIRAFFDGKPLRAYRPPTRRWGSVLGI-QPDGLEVFAPNGRVFRF
Mvan_5760|M.vanbaalenii_PYR-1 GLLARNIRAAFAGKPLREYRAPKRRWGSVLGI-QPDGLEVFAPNGRAFRF
MSMEG_6582|M.smegmatis_MC2_155 ALVAHNVAAYFTGGPMRTYRPHGRRWGSVLGP-QPDGLDVFAPNGFAFHF
Mb1842c|M.bovis_AF2122/97 RYAGCNVINDLFDQPLLALRIPWYVTVLDLG---SAGAVYTEGWERKVVS
Rv1812c|M.tuberculosis_H37Rv RYAGCNVINDLFDQPLLALRIPWYVTVLDLG---SAGAVYTEGWERKVVS
MLBr_02061|M.leprae_Br4923 RFSAVAKIGPLEFSGFIAWLMWLVLHLMYLIG-FKTKITTLLSWTVTFLS
MAB_2429c|M.abscessus_ATCC_199 RYSAVVKIGKIEFSGFIAWLSWLALHLVYLVG-FKNRLVTLILWSIAFVT
TH_4200|M.thermoresistible__bu SFGLTEEQAREEGYDVKVAKFPFTANGKAHGMGAPSGFVKLIADAKYGEL
.
MMAR_2587|M.marinum_M PSWSVHRVVMPVVVRWGMYRGVRKNHPLT---------------------
MUL_3173|M.ulcerans_Agy99 PSWSVHRVVMPVVVRWGMYRGVRKNHPLT---------------------
MAV_3047|M.avium_104 PSWSVQRVVMPLIVRWGMYGGVRRNDPLGRETATPPTL------------
Mflv_1066|M.gilvum_PYR-GCK PAWAFDRVLMPVIVRWGIYRGVRAGGSRR---------------------
Mvan_5760|M.vanbaalenii_PYR-1 PVWAFDRVLMPGIVRWGIYRGVRQDVS-----------------------
MSMEG_6582|M.smegmatis_MC2_155 PGWSIQRVLQPWIVRRVIYRGLRDPV------------------------
Mb1842c|M.bovis_AF2122/97 QG-APAKTTKQSINTRRIYPPLNGSRADLLAAAAPRVQPRP---------
Rv1812c|M.tuberculosis_H37Rv QG-APAKTTKQSINTRRIYPPLNGSRADLLAAAAPRVQPRP---------
MLBr_02061|M.leprae_Br4923 TRRGQLTITEQQAFARTRLEQLAELTAESQSTAAANRATMR---AS----
MAB_2429c|M.abscessus_ATCC_199 RSRGQMAITEQQAWARTRLDQLEELVAEKNGDEEPTTQPDKEKAAS----
TH_4200|M.thermoresistible__bu LGGHMVGHDVSELLPELTLAQKWDLTAHELARNVHTHPTLSEAMQECFHG
MMAR_2587|M.marinum_M --------
MUL_3173|M.ulcerans_Agy99 --------
MAV_3047|M.avium_104 --------
Mflv_1066|M.gilvum_PYR-GCK --------
Mvan_5760|M.vanbaalenii_PYR-1 --------
MSMEG_6582|M.smegmatis_MC2_155 --------
Mb1842c|M.bovis_AF2122/97 --------
Rv1812c|M.tuberculosis_H37Rv --------
MLBr_02061|M.leprae_Br4923 --------
MAB_2429c|M.abscessus_ATCC_199 --------
TH_4200|M.thermoresistible__bu LVGHMINF