For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VFTEAMTAERIARSDRSTGTREAILSAAEVLFAERGMYAVSNRQISEAAGQGNNAAACYHFGTRVDLLRA IEGKHREPIEKLRAQMLAAVGDSTELRDWVGTLVRPLTDHLSALGTPSWYARFAAQAMADPSYRHVVTKD ALSSPLLVRTLDGINRCLPELPRRVRAERMVMVRNLLMHTCAEHEGALAEHGPRSRSAWPVAAEGLIDAI VGLWRAPVHVAAAAGPTGQTTAPATGQTDHRGAP
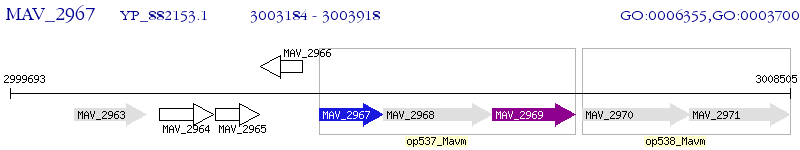
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2967 | - | - | 100% (244) | transcriptional regulator, TetR family protein |
| M. avium 104 | MAV_0887 | - | 6e-74 | 62.56% (219) | transcriptional regulator, TetR family protein |
| M. avium 104 | MAV_2626 | - | 3e-16 | 30.45% (220) | transcriptional regulator, TetR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2416 | - | 2e-80 | 65.44% (217) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0835c | - | 5e-11 | 38.05% (113) | TetR family transcriptional regulator |
| M. marinum M | MMAR_4764 | - | 8e-73 | 63.68% (212) | hypothetical protein MMAR_4764 |
| M. smegmatis MC2 155 | MSMEG_4858 | - | 1e-82 | 67.13% (216) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2278 | - | 8e-80 | 65.44% (217) | transcriptional regulator, TetR family protein |
| M. ulcerans Agy99 | MUL_0335 | - | 4e-72 | 63.68% (212) | hypothetical protein MUL_0335 |
| M. vanbaalenii PYR-1 | Mvan_4236 | - | 4e-81 | 65.90% (217) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2416|M.gilvum_PYR-GCK MQSGATTFLNGTTHLKQSLDLPGVTRFTDRVTTARTVRADRATGTQEAIL
Mvan_4236|M.vanbaalenii_PYR-1 -----MSFLN------QSLDLVGVTGFTEWVTTARTVRADRASSTQEAIL
MMAR_4764|M.marinum_M -----------------------------MTAVGRAVRGERASSTQEAIL
MUL_0335|M.ulcerans_Agy99 -----------------------------MTAVGRAVRGERASSTQEAIL
MSMEG_4858|M.smegmatis_MC2_155 --------------MTRFTGLVTTASRTASRTAARTARADRATSTQEAIL
TH_2278|M.thermoresistible__bu -----------------------VVQFTEGVTAARTVRADRASSTQEAIL
MAV_2967|M.avium_104 -------------------------MFTEAMTAERIARSDRSTGTREAIL
MAB_0835c|M.abscessus_ATCC_199 -----------------------------MTDTSAETTILTEDSTDQRLL
. . .* : :*
Mflv_2416|M.gilvum_PYR-GCK KAAERLYAEHGVFAVSNRQVSEAAGQGNNAAVGYHFGTKADLVRAIEHKH
Mvan_4236|M.vanbaalenii_PYR-1 KAAERLYAEHGVFAVSNRQVSEAAGQGNNAAVGYHFGTKADLVRAIEHKH
MMAR_4764|M.marinum_M VAAERLFAEHGVFAVSNRQVSEAAGQGNNAAVGYHFGTKTDLVRAIEQKH
MUL_0335|M.ulcerans_Agy99 VAAERLFAEHGVFAVSNRQVSEAAGQGNNAAVGYHFGTKTDLVRAIEQKH
MSMEG_4858|M.smegmatis_MC2_155 KAAERLYAEHGVFAVSNRQVSEAAGQGNNAAVGYHFGTKTDLVRAIEHKH
TH_2278|M.thermoresistible__bu KAAERLYAERGMFAVSNRQVSEAAGQGNNAAVGYHFGTKTDLVRAIEQKH
MAV_2967|M.avium_104 SAAEVLFAERGMYAVSNRQISEAAGQGNNAAACYHFGTRVDLLRAIEGKH
MAB_0835c|M.abscessus_ATCC_199 NAAERLFAERGVDAVSLRAIMAAAG-TNVASVHYHFGSKERLIEALLDRY
*** *:**:*: *** * : *** * *:. ****:: *:.*: ::
Mflv_2416|M.gilvum_PYR-GCK RGPIEELREQRVAAIDSS---GDMRDWVAALVCPLTDHLAALGNPTWYAR
Mvan_4236|M.vanbaalenii_PYR-1 RGPIEELREQRVAAIGAS---TDMRDWVAALVCPLTDHLAALGNPTWYAR
MMAR_4764|M.marinum_M RVPVERLREQMVAAAAAKGVAATMRDWVACLVCPLTEHLAVLGNPTWYAR
MUL_0335|M.ulcerans_Agy99 RVPIERLREQMVAAAAAKGAAATMRDWVACLVCPLTEHLAVLGNPTWYAR
MSMEG_4858|M.smegmatis_MC2_155 RGPIEYLREQMIGTVQGS---TDLRDWVGALVRPLTDHLAALGNPTWYAR
TH_2278|M.thermoresistible__bu RVEIERLLEKMVAETGDS---TDLRDWVACLVCSLTEHLHQLGNPTWYAR
MAV_2967|M.avium_104 REPIEKLRAQMLAAVGDS---TELRDWVGTLVRPLTDHLSALGTPSWYAR
MAB_0835c|M.abscessus_ATCC_199 LDQIEKRRFALLDVAETA---RTLRAIAEAIVTPLS-EIAEEGSTAWLST
:* : :* . :* .*: .: *..:* :
Mflv_2416|M.gilvum_PYR-GCK FAAQVMT-DPAYHNMIVKDALSSPSLVRVLDGINGCLPDLPVDVRVERNI
Mvan_4236|M.vanbaalenii_PYR-1 FAAQVMT-DPAYHNMIVKDALSSPSLVRVLDGINSCLPDLPVDVRVERNI
MMAR_4764|M.marinum_M FAAQAMT-DPAYHNIIVKDALSSPSLVQVIDGINSCLPDLPVDVRYERNL
MUL_0335|M.ulcerans_Agy99 FAAQAMT-DPAYHNIIVKDALSSPSLVQVIDGINSCLPDLPVDVRYERNL
MSMEG_4858|M.smegmatis_MC2_155 FAAQVMT-DPAYHNIIVRDALSSPSLVQVIEGMNNCLPDLPAEIRAERNI
TH_2278|M.thermoresistible__bu FAAQALA-DPAYQKIVVKDALASPALVRVVEGITRCLPDLPAAVVVERNI
MAV_2967|M.avium_104 FAAQAMA-DPSYRHVVTKDALSSPLLVRTLDGINRCLPELPRRVRAERMV
MAB_0835c|M.abscessus_ATCC_199 LGKLLISGHPALIPMSVSFQPQAARLQELILRLRPDATLPSIRFGVTHAV
:. :: .*: : . :. * . : : . . . : :
Mflv_2416|M.gilvum_PYR-GCK MGRNLLMHSCADRERLLAQGSPVPRGSWQSAATGLIDAIIGLWHAPVTER
Mvan_4236|M.vanbaalenii_PYR-1 MGRNLLMHSCADRERLLAQGAPVARPSWQAAATGLIDAIIGLWHAPVTEP
MMAR_4764|M.marinum_M MARNLLMHTCADLERALAAGTSVPRPSWREAATGLIDAITGLWLAPVTAR
MUL_0335|M.ulcerans_Agy99 MARNLLMHTCADLERALAAGTSVPRPFWREAATGQIDAITGLWLAPVTAR
MSMEG_4858|M.smegmatis_MC2_155 MARNLLMHSCAERERALAQGAPVPRSSWQGAASGLIDAIVGLWLAPVTGE
TH_2278|M.thermoresistible__bu MARNLLMHTCADFERAFAEGDPVPRASWQAVGNGLIDAIVGLWRAPVTEP
MAV_2967|M.avium_104 MVRNLLMHTCAEHEGALAEHGPRSRSAWPVAAEGLIDAIVGLWRAPVHVA
MAB_0835c|M.abscessus_ATCC_199 TLTCVVLGDIAHTNRLMSLSGTYLDD--GQMNAQLIDMVTAILAGPPDES
::: *. : :: . ** : .: .*
Mflv_2416|M.gilvum_PYR-GCK P----------------------
Mvan_4236|M.vanbaalenii_PYR-1 A----------------------
MMAR_4764|M.marinum_M E----------------------
MUL_0335|M.ulcerans_Agy99 E----------------------
MSMEG_4858|M.smegmatis_MC2_155 DR---------------------
TH_2278|M.thermoresistible__bu PQ---------------------
MAV_2967|M.avium_104 AAAGPTGQTTAPATGQTDHRGAP
MAB_0835c|M.abscessus_ATCC_199 -----------------------