For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDLNETRERIIYQKEGPIARVTLNWPEKANAQDQKLAEEVDAALADADRDYDIKVLIMKANGKGFCSGHA IGNNAVDYPAFVEGAKVMGTPWKPQTDLFVKPILNLWEFSKPTIAQVHGYCVGGGTHYGLTTDIVIAAED AYFSYPPLQGFGMPSGECSIEPWIFMNWRRAAYYLYLAEVIDARKALEVGLVNEVVPRDQLDERVEAIAR HIAQAPMTTLLATKANLKRAWELMGMRVHWQSSNDLVALASISKDVQELIQTVFKDKVLPSEQARRQAAA AAQTDGAATT
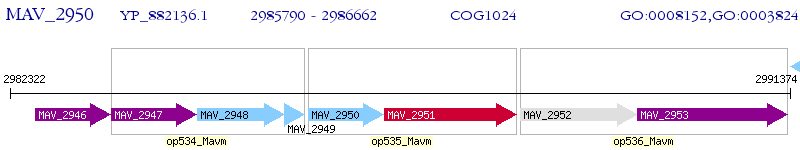
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2950 | - | - | 100% (290) | enoyl-CoA hydratase/isomerase family protein |
| M. avium 104 | MAV_2954 | - | 4e-72 | 50.94% (267) | enoyl-CoA hydratase/isomerase family protein |
| M. avium 104 | MAV_1848 | - | 5e-72 | 48.29% (263) | enoyl-CoA hydratase/isomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0464c | echA2 | 1e-32 | 30.94% (265) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2433 | - | 8e-26 | 32.35% (238) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv0456c | echA2 | 1e-32 | 30.94% (265) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_00120 | echA1 | 1e-14 | 28.21% (234) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_3859c | - | 8e-25 | 30.54% (239) | enoyl-CoA hydratase |
| M. marinum M | MMAR_0778 | echA2 | 3e-32 | 29.29% (280) | enoyl-CoA hydratase EchA2 |
| M. smegmatis MC2 155 | MSMEG_1388 | - | 5e-28 | 32.91% (234) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_3498 | - | 2e-77 | 51.12% (268) | PUTATIVE enoyl-CoA hydratase/isomerase |
| M. ulcerans Agy99 | MUL_1414 | echA2 | 2e-32 | 29.29% (280) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_0780 | - | 5e-32 | 30.00% (280) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2950|M.avium_104 ------MDLNETRERIIYQKEGPIARVTLNWPEKANAQDQKLAEEVDAAL
TH_3498|M.thermoresistible__bu ------LET-RTLEHVIYEKDGPIARIILNNPDRANAQTSEMVHSVNTAL
Mb0464c|M.bovis_AF2122/97 -------MPTPDFQTLLYTTAGPVATITLNRPEQLNTIVPPMPDEIEAAI
Rv0456c|M.tuberculosis_H37Rv -------MPTPDFQTLLYTTAGPVATITLNRPEQLNTIVPPMPDEIEAAI
MMAR_0778|M.marinum_M -------MPTPEFQTLLYSTAGPVATITLNRPEHLNTIVPPMPDEIEAAV
MUL_1414|M.ulcerans_Agy99 -------MPTPEFQTLLYSTAGPVATITLNRPEHLNTIVPPMPDEIEAAV
Mvan_0780|M.vanbaalenii_PYR-1 -------MPEPEFETLLYRADSAVATITLNRPEQLNTIVPPMPDEIEAAV
MAB_3859c|M.abscessus_ATCC_199 --------MVDTLKTMTYEVTDRVARITFNRPEQGNAIIADTPLELAALV
MSMEG_1388|M.smegmatis_MC2_155 MTHAIRPVDFDNLKTMTYEVTDRVARITFNRPEKGNAIVADTPLELSALV
Mflv_2433|M.gilvum_PYR-GCK -------------MYIDYEVSDRIATITLNRPEAANAQNPELLDELDAAW
MLBr_00120|M.leprae_Br4923 ----MGSDINRAYESVAVEIKDRVAQVTLIGPGKGNAMGPAFWSEMPEVF
: . :* : : * *: .:
MAV_2950|M.avium_104 ADADRDYDIKVLIMKANGKGFCSGHAIGNN-------AVDYPAFVEGAK-
TH_3498|M.thermoresistible__bu DDAQYDYDIKVVIIKANGKGFCSGHIPDG----------SYPEFKAEIE-
Mb0464c|M.bovis_AF2122/97 GLAERDQDIKVIVLRGAGRAFSGGYDF----------GGGFQHWGDAMM-
Rv0456c|M.tuberculosis_H37Rv GLAERDQDIKVIVLRGAGRAFSGGYDF----------GGGFQHWGDAMM-
MMAR_0778|M.marinum_M GLAERDQGIKVIVLRGAGRAFSGGYDF----------GGGFQHWGETMM-
MUL_1414|M.ulcerans_Agy99 GLAERDQGIKVIVLRGAGRAFSGGYDF----------GGGFQHWGETMM-
Mvan_0780|M.vanbaalenii_PYR-1 GLAERDPQVKVIVLRGAGRAFSGGYDF----------GGGFAHWGEAMN-
MAB_3859c|M.abscessus_ATCC_199 ERADLDPQVHVILVSGRGDGFCGGFDLSAYAEAND--EGSVRYSGTALDG
MSMEG_1388|M.smegmatis_MC2_155 ERADLDPDVHVILVSGRGEGFCAGFDLSAYAEGSSSAGGGSPYEGTVLSG
Mflv_2433|M.gilvum_PYR-GCK TRAAEDNDVSVIVLRANGKHFSAGHDLRG---------------------
MLBr_00120|M.leprae_Br4923 AELDTNREVRAVVITGSGKDFSYGLDVPAMG----------GKFVPLLT-
: : .::: . * *. *
MAV_2950|M.avium_104 -------VMGTPWKPQTDLFVKP---------ILNLWEFSKPTIAQVHGY
TH_3498|M.thermoresistible__bu -------ASGKVWRSAAQLFLWP---------VLKLWEFPKPVIAQVHGY
Mb0464c|M.bovis_AF2122/97 --------TDGRWDPGKDFAMVTARETGPTQKFMAIWRASKPVIAQVHGW
Rv0456c|M.tuberculosis_H37Rv --------TDGRWDPGKDFAMVTARETGPTQKFMAIWRASKPVIAQVHGW
MMAR_0778|M.marinum_M --------TDGQWDPGKDFAMVTARETAPTQKFMAIWRASKPVIAQIHGW
MUL_1414|M.ulcerans_Agy99 --------TDGQWDPGKDFAMVTARETAPTQKFMAIWRASKPVIAQIHGW
Mvan_0780|M.vanbaalenii_PYR-1 --------TDGRWDPGKDFAMVSARETGPTQKFMAIWRASKPVIAQVHGW
MAB_3859c|M.abscessus_ATCC_199 QVQLRNHLPNINWDPMLDYQMMSRFTRG----FSSLLHANKPTVVKVHGY
MSMEG_1388|M.smegmatis_MC2_155 KTQALNHLPDEPWDPMVDYQMMSRFVRG----FASLMHCDKPTVVKIHGY
Mflv_2433|M.gilvum_PYR-GCK --------GGPVPDKITLEFIIQHEAKRYLEYTLRWRNVPKPSIAAVQGR
MLBr_00120|M.leprae_Br4923 --------DGALARPRTDFHAEVLRMQK---AINAVADCRTPTIAAVHGW
. .* :. ::*
MAV_2950|M.avium_104 CVGGGTHYGLTTDIVIAAEDAYFSYPPLQGFGMPSGECSIEPWIFMNWRR
TH_3498|M.thermoresistible__bu AIGGGTTWALIPEITVCSEDAWFQMPLVPGFGLPGSETMFEPWVFMNYKR
Mb0464c|M.bovis_AF2122/97 CVGGASDYALCADIVIASEDAVIGTPYSRMWGAYLTGMWLYRLSLA---K
Rv0456c|M.tuberculosis_H37Rv CVGGASDYALCADIVIASEDAVIGTPYSRMWGAYLTGMWLYRLSLA---K
MMAR_0778|M.marinum_M CVGGASDYALCADIVIASDDAVIGTPYSRMWGAYLSGMWLYRLSFA---K
MUL_1414|M.ulcerans_Agy99 CVGGASDYALCADIVIASDDAVIGTPYSRMWGAYLSGMWLYRLSFA---K
Mvan_0780|M.vanbaalenii_PYR-1 CVGGASDYALCADIVIASDDAVIGTPYARMWGAYLTGMWIYRLSLA---K
MAB_3859c|M.abscessus_ATCC_199 CVAGGTDIALHADQLIVAADAKIGYPPTRVWGVPATGLWAHKLGDQ---R
MSMEG_1388|M.smegmatis_MC2_155 CVAGGTDIALHADQVIAAADAKIGYPPMRVWGVPAAGLWAHRLGDQ---R
Mflv_2433|M.gilvum_PYR-GCK CISGGLLLCWPCDLIVAADDAQFSDP-VVLMGIGGVEYHGHTWELGP-RK
MLBr_00120|M.leprae_Br4923 CIGGALDLISAVDIRYASADAKFSMREVKLAIVADMGSLARLPLILSDGH
.:.*. : : ** : :
MAV_2950|M.avium_104 AAYYLYLAEVIDARKALEVGLVNEVVP-RDQLDERVEAIARHIAQAPMTT
TH_3498|M.thermoresistible__bu AAEYLYTAQKITAQQALEFGLVNRVVP-ADELESTVEELAAKIAKAPLIT
Mb0464c|M.bovis_AF2122/97 VKWHSLTGRPLTGVQAAEAELINEAVP-FERLEARVAEIATELARIPLSQ
Rv0456c|M.tuberculosis_H37Rv VKWHSLTGRPLTGVQAAEAELINEAVP-FERLEARVAEIATELARIPLSQ
MMAR_0778|M.marinum_M AKWHSLTGRPLSGVQAAQIELINEAVP-FERLEARVAEVAAELAQIPLSQ
MUL_1414|M.ulcerans_Agy99 AKWHSLTGRPLSGVQAAQIELINEAVP-FERLEARVAEVAAELAQIPLSQ
Mvan_0780|M.vanbaalenii_PYR-1 VKWHSLTGEPLTGKEAAAVELINESVP-FEQLEARVAEVAQKLAGIPLSQ
MAB_3859c|M.abscessus_ATCC_199 AKRLLLTGDCLSGRQAYDWGLAVEAPE-PDELDERTERLVERIAAMPLNQ
MSMEG_1388|M.smegmatis_MC2_155 AKRLLFTGDCITGAQAAEWGLAVEAPD-PADLDARTERLVERIAAMPVNQ
Mflv_2433|M.gilvum_PYR-GCK AKEILFTGRAMTADEVAATGMVNKVVP-RAELDAETRALAEQIAKMPPFA
MLBr_00120|M.leprae_Br4923 LRELALTGKDIDATRAEKIGLVNDVYDNADKTLAAAHATAAEIAANPPLA
. : . .. : . . .:* *
MAV_2950|M.avium_104 LLATKANLKRAWELMGMRVHWQSSNDLVALASISKDVQELIQTVFKDKVL
TH_3498|M.thermoresistible__bu LQATKAGILRAWETMGFRTHQQASNDLQAVVTGSREFQEYMAELMKRASK
Mb0464c|M.bovis_AF2122/97 LQAQKLIVNQAYENMGLASTQLLGGILDGLMRNTPDALEFIRTAQTQGVR
Rv0456c|M.tuberculosis_H37Rv LQAQKLIVNQAYENMGLASTQLLGGILDGLMRNTPDALEFIRTAQTQGVR
MMAR_0778|M.marinum_M LQAQKLIVNQAYENMGLASTQTLGGILDGLMRNTPDALDFIKTAENQGVR
MUL_1414|M.ulcerans_Agy99 LQAQKLIVNQAYENMGLASTQTLGGILDGLMRNTPDALDFIKTAENQGVR
Mvan_0780|M.vanbaalenii_PYR-1 LQAQKLIVNQAFENMGLASTQTLGGILDGLMRNTPDALGFIDTAAADGVR
MAB_3859c|M.abscessus_ATCC_199 LMMIKLACNSVLINQGIANSAMIGTVFDGISRHTPEAHAFTADAIANGYR
MSMEG_1388|M.smegmatis_MC2_155 LIMAKLACNTALLNQGVATSQMVSTVFDGIARHTPEGHAFVATAREHGFR
Mflv_2433|M.gilvum_PYR-GCK LRQAKRAVNQTLDVQGFYAAIQSVFDVHQTGHGNALSVGGWPVLVNLEEM
MLBr_00120|M.leprae_Br4923 VYGVKDVLYQQRT-SAISESLRYVAAWNAAFLPSKDLTEGISATFAKRAP
: * .. .
MAV_2950|M.avium_104 PSEQARRQAAAAAQTDGAATT-------------
TH_3498|M.thermoresistible__bu PADRV-----------------------------
Mb0464c|M.bovis_AF2122/97 AAVERRDGPFGDYSQAPPELRPDPTHVITPDGSM
Rv0456c|M.tuberculosis_H37Rv AAVERRDGPFGDYSQAPPELRPDPTHVITPDGSM
MMAR_0778|M.marinum_M AAVERRDGPFGDYSQAPPELRPNPNHVIVPDRDR
MUL_1414|M.ulcerans_Agy99 AAVERRDGPFGDYSQAPPELRPNPNHVIVPDRDR
Mvan_0780|M.vanbaalenii_PYR-1 AAVEQRDGPWGDYSQAPPHRRPDPSHVIEP----
MAB_3859c|M.abscessus_ATCC_199 QAVRHRDEPFGDYGRKPTEL--------------
MSMEG_1388|M.smegmatis_MC2_155 EAVRRRDEPMGDHGRRASDV--------------
Mflv_2433|M.gilvum_PYR-GCK KANIK-----------------------------
MLBr_00120|M.leprae_Br4923 QFIGE-----------------------------