For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNVTVTVVSIVAIAVLIFGNAVFVAAEFSLTTLDRSTVEANARTGGRRDRWIRRAHHRLSFQLSGAQLGI SATTLAAGYLTDPMVEDLPHPWLDAVGIPDRVGDAIMAFLALLIVTSVSMVFGELVPKYLAVARPMSTAR AVVAAQLLFSALLTPAIRLTNGAANWIVRRLGVEPAEELRSARSPQELLSLVRSSARSGALDAATAALVR RSLQFGTLSAEELMTPRSKIVALQTDDTVADLVTAAAESGFSRFPIVDGDLDETVGIVHVKQVFAIPRAA RAHTLLTTVAQPVPVVPSTLDGDAVMAEIRANSLQTALVVDEYGGTAGMVTVEDLIEEIVGDVRDEHDDA TPDVVPAGTGWRVSGLLRIDEVAAATGFRAPEGPYETIGGLVLREIGHIPAAGETVKLPALDPDGLLDPV LRWQARVIRMDGRRIDVLELTEIDAHAEAPGGRR
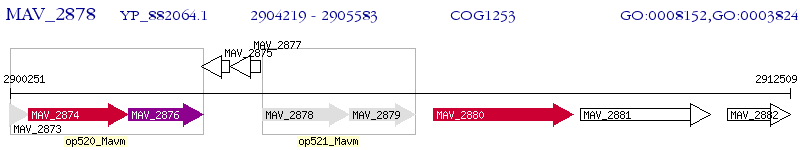
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2878 | - | - | 100% (454) | CBS domain-containing protein |
| M. avium 104 | MAV_2879 | - | 9e-41 | 32.55% (341) | CBS domain-containing protein |
| M. avium 104 | MAV_2028 | - | 2e-32 | 30.92% (304) | CBS domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1873c | - | 0.0 | 78.81% (453) | hypothetical protein Mb1873c |
| M. gilvum PYR-GCK | Mflv_3364 | - | 1e-167 | 66.15% (452) | hypothetical protein Mflv_3364 |
| M. tuberculosis H37Rv | Rv1842c | - | 0.0 | 78.81% (453) | hypothetical protein Rv1842c |
| M. leprae Br4923 | MLBr_00387 | guaB2 | 5e-06 | 23.64% (165) | inosine 5'-monophosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2411c | - | 1e-161 | 66.37% (443) | hypothetical protein MAB_2411c |
| M. marinum M | MMAR_2716 | - | 0.0 | 79.01% (443) | hypothetical protein MMAR_2716 |
| M. smegmatis MC2 155 | MSMEG_3637 | - | 1e-174 | 69.25% (452) | CBS domain-containing protein |
| M. thermoresistible (build 8) | TH_2608 | - | 1e-169 | 68.85% (443) | PUTATIVE protein of unknown function DUF21 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3094 | - | 1e-168 | 68.07% (451) | hypothetical protein Mvan_3094 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1873c|M.bovis_AF2122/97 -----MNLT---DTVATILAILALTAG---------------TGVFVAAE
Rv1842c|M.tuberculosis_H37Rv -----MNLT---DTVATILAILALTAG---------------TGVFVAAE
MMAR_2716|M.marinum_M -----MMFT---ATTVSIVAILVLTLG---------------TAVFVGAE
MAV_2878|M.avium_104 -----MNVT---VTVVSIVAIAVLIFG---------------NAVFVAAE
Mflv_3364|M.gilvum_PYR-GCK -----MNTA---MSLLALLAFMLLTAG---------------TAVFVAAE
Mvan_3094|M.vanbaalenii_PYR-1 -----MSSN---LSLLALLAFMLLTAG---------------TAVFVAAE
TH_2608|M.thermoresistible__bu -----MSSV---SVALSILAILLLTAG---------------TAAFVAAE
MSMEG_3637|M.smegmatis_MC2_155 MGEVAMSDTNVWLSLLAILAVVLLTFG---------------TAVFVAAE
MAB_2411c|M.abscessus_ATCC_199 -----MTVL---LTGLSLLAFVALTAG---------------TALFVAAE
MLBr_00387|M.leprae_Br4923 ----MIRGMSNLKESSDFVASSYVRLGGLMDDPAATGGDNPHKVAMLGLT
: ::* : * . ::.
Mb1873c|M.bovis_AF2122/97 FSLTALDRSTVEANARGGTSRDRFIQRAHHRLSFQLSGAQLG----ISIT
Rv1842c|M.tuberculosis_H37Rv FSLTALDRSTVEANARGGTSRDRFIQRAHHRLSFQLSGAQLG----ISIT
MMAR_2716|M.marinum_M FSLTALDRSTVEANARGGDSRDRFIRRAQHRLSFHLSGAQLG----ISIT
MAV_2878|M.avium_104 FSLTTLDRSTVEANARTGGRRDRWIRRAHHRLSFQLSGAQLG----ISAT
Mflv_3364|M.gilvum_PYR-GCK FSLTALERSTVEANVRSGDRRDAMVQRAHRSLSTQLSGAQVG----ISIT
Mvan_3094|M.vanbaalenii_PYR-1 FSLTALERSTVEANVRSGDRRDVMVQRAHRTLSTQLSGAQVG----ISIT
TH_2608|M.thermoresistible__bu FSLTALERSTVEANARSGDRRDQMVRRAHRTLSFQLSGAQVG----ISIT
MSMEG_3637|M.smegmatis_MC2_155 FSLTALERSTVEANARAGDRRDHMVRHAHRTLSFQLSGAQVG----ISIT
MAB_2411c|M.abscessus_ATCC_199 FSLTALERSGIEARAKTGDSRDRLVKRAHSTLSFQLSGAQLG----ITIT
MLBr_00387|M.leprae_Br4923 FDDVLLLPAASDVVPATADISSQLTKKIRLKVPLVSSAMDTVTEARMAIA
*. . * : :. . . :: : :. *. : :: :
Mb1873c|M.bovis_AF2122/97 TLATGYLTEPLVAELPHPGLVAVGMSDRVADGLITFFALVIVTSLSMVFG
Rv1842c|M.tuberculosis_H37Rv TLATGYLTEPLVAELPHPGLVAVGMSDRVADGLITFFALVIVTSLSMVFG
MMAR_2716|M.marinum_M TLATGYLTEPLVDALPHPALTAIGISDQMSDAITVFLALVVVTSVSMVFG
MAV_2878|M.avium_104 TLAAGYLTDPMVEDLPHPWLDAVGIPDRVGDAIMAFLALLIVTSVSMVFG
Mflv_3364|M.gilvum_PYR-GCK TLATGFLAEPVVARLIHPGLTALNVPGQFVSGLSLFLAILIATSISMIFG
Mvan_3094|M.vanbaalenii_PYR-1 TLATGFLAEPVVARLIHPGLTAIGIPDRFVGGLALTLAILIATSISMVFG
TH_2608|M.thermoresistible__bu TLATGYLAEPVVARLLQPAIDAVGVPDRLAGGLSLFLAVSIATSLSMVFG
MSMEG_3637|M.smegmatis_MC2_155 TLATGYLAEPVIGKLIHPGLDAIGIPANFAGGLSLFFAVVIATSLSMVFG
MAB_2411c|M.abscessus_ATCC_199 TLVTGYLAEPVIARLLSQLFGYTGIA--VSSAISLALALIIATSISMIFG
MLBr_00387|M.leprae_Br4923 MARAGGMGVLHRNLPVGEQAGQVETVKRSEAGMVTDPVTCRPDNTLAQVG
:* : .: . . .*
Mb1873c|M.bovis_AF2122/97 ELVPKYLAVARPLRTARSVVA-----GQVLFSLLLTPAIRLTNGAANWIV
Rv1842c|M.tuberculosis_H37Rv ELVPKYLAVARPLRTARSVVA-----GQVLFSLLLTPAIRLTNGAANWIV
MMAR_2716|M.marinum_M ELVPKYLAVARPLPTARAVVL-----PQWLFSVLLTPAIRLTNGAANWIV
MAV_2878|M.avium_104 ELVPKYLAVARPMSTARAVVA-----AQLLFSALLTPAIRLTNGAANWIV
Mflv_3364|M.gilvum_PYR-GCK ELVPKNLAVAKPVPTARWSAP-----LQLMFSFLALPLIRLTNGTANWIL
Mvan_3094|M.vanbaalenii_PYR-1 ELVPKNLAVARPVPTARWSAP-----LQLMFSFLFTPLIRLTNGTANWIL
TH_2608|M.thermoresistible__bu ELVPKNLAVARPVETARWSAP-----LQVVFSALVTPLIRVTNGTANWIL
MSMEG_3637|M.smegmatis_MC2_155 ELVPKNLAVARPVPTARAAAP-----FQLLFSLIFTPVIRLTNGTANWIL
MAB_2411c|M.abscessus_ATCC_199 ELVPKNFALARPVWTSRATSG-----PMVLFSAVFAFAIRALNGTANWVL
MLBr_00387|M.leprae_Br4923 ALCARFRISGLPVVDDSGALAGIITNRDMRFEVDQSKQVAEVMTKTPLIT
* .: . *: *. : : :
Mb1873c|M.bovis_AF2122/97 RRLGIEPAEELRSAR-TPQELVSLVRSSARSGALDDATAWLMRRSLQFGA
Rv1842c|M.tuberculosis_H37Rv RRLGIEPAEELRSAR-TPQELVSLVRSSARSGALDDATAWLMRRSLQFGA
MMAR_2716|M.marinum_M RQLGIEPAEELRSAR-TPQELVSLVRSSARSGSLDDVTASLMRRSLQFGT
MAV_2878|M.avium_104 RRLGVEPAEELRSAR-SPQELLSLVRSSARSGALDAATAALVRRSLQFGT
Mflv_3364|M.gilvum_PYR-GCK RRLGIEPAEELRSAR-SPQELVSLVRSSAERGSLDPVTAQLVDRSLQFGD
Mvan_3094|M.vanbaalenii_PYR-1 RRLGIEPAEELRSAR-SPQELVSLVRSSAERGSLDPVTALLVDRSLQFGD
TH_2608|M.thermoresistible__bu RRLGIEPAEELRSAR-SAQELVSLVRNSAQSGSLDPVTARLVDRSLQFGD
MSMEG_3637|M.smegmatis_MC2_155 RRLGIEPAEELRSAR-TGQELAALVRNSARSGSLDPVTALLVDRSLQFGE
MAB_2411c|M.abscessus_ATCC_199 RRMGIEPAEELRSAR-SPQELGSLVRTSARSGALDETTAVLVDRSLRFGS
MLBr_00387|M.leprae_Br4923 AAEGVSADAALGLLRRNKIEKLPVVDGHGRLTGLITVKDFVKTEQHPLAT
*:.. * * . * .:* .. .* .. : .. :.
Mb1873c|M.bovis_AF2122/97 LTAEELMTP--------RSKIVALQTDDTIADLVAAAAASGFSRFPV-VE
Rv1842c|M.tuberculosis_H37Rv LTAEELMTP--------RSKIVALQTDDTIADLVAAAAASGFSRFPV-VE
MMAR_2716|M.marinum_M LTAEELMTP--------RSKVVALQTDDTIADLVAAVAASGYSRFPV-VQ
MAV_2878|M.avium_104 LSAEELMTP--------RSKIVALQTDDTVADLVTAAAESGFSRFPI-VD
Mflv_3364|M.gilvum_PYR-GCK RTAEELMTP--------RSKIDTLDADDTVLDLSEAAVSTGHSRFPV-IH
Mvan_3094|M.vanbaalenii_PYR-1 RSAEELMTP--------RSKIDTLEADDTVADLSDAATRTGHSRFPV-IR
TH_2608|M.thermoresistible__bu RTAEELMTP--------RSKIEALDADDTVADLIRVADETGFSRFPI-TR
MSMEG_3637|M.smegmatis_MC2_155 RTAEELMTP--------RPKIETLDADDTVVDLMDAAIRTGFSRFPI-IK
MAB_2411c|M.abscessus_ATCC_199 RTAEELMTP--------RTEIESLEATDTVADLVTIAVETGFSRFPI-TE
MLBr_00387|M.leprae_Br4923 KDNDGRLLVGAAVGVGGDAWVRAMMLVDAGVDVLIVDTAHAHNRLVLDMV
: : . : :: *: *: ...*: :
Mb1873c|M.bovis_AF2122/97 GDLDATVG--IVHVKQVFEVPPGDRAHTLLTTVAEPVAVVPSTLDGDAVM
Rv1842c|M.tuberculosis_H37Rv GDLDATVG--IVHVKQVFEVPPGDRAHTLLTTVAEPVAVVPSTLDGDAVM
MMAR_2716|M.marinum_M GDLDETFG--IVHVKQVFQVPAAKRAHTLLSSIAKPVPVVPSTLDGDALM
MAV_2878|M.avium_104 GDLDETVG--IVHVKQVFAIPRAARAHTLLTTVAQPVPVVPSTLDGDAVM
Mflv_3364|M.gilvum_PYR-GCK GDLDETIG--MVHVKQVFEVPAVSRSTTRLADLVQPLTKVPSTLDGDAVM
Mvan_3094|M.vanbaalenii_PYR-1 GDLDETVG--MVHVKQVFAVPADARATTRLATLVQPVTKVPSTLDGDAVM
TH_2608|M.thermoresistible__bu GDLDETVG--MVHVKQVFEVPHDQRATTRLARLAVPVIKVPSTLDGDALM
MSMEG_3637|M.smegmatis_MC2_155 GDLDETIG--VVHVKQVFAVPRDKRDRVRLASLALPVAKVPSTLDGDAVM
MAB_2411c|M.abscessus_ATCC_199 GDLDATIG--IVHVKQVFEVPRDRRATTTLASLARPVAVVPSTLDGDAVM
MLBr_00387|M.leprae_Br4923 GKLKVEIGDRVQVIGGNVATRSAAAALVEAGADAVKVGVGPGSTCTTRVV
*.*. .* : : . . . : *.: ::
Mb1873c|M.bovis_AF2122/97 AQVRASALQTAMVVDEYGGTAGMVTLEDLIEEIVGDVRDEHDDATPDVVA
Rv1842c|M.tuberculosis_H37Rv AQVRASALQTAMVVDEYGGTAGMVTLEDLIEEIVGDVRDEHDDATPDVVA
MMAR_2716|M.marinum_M AQIRANSLQTAMVVDEYGGTAGMVTVEDLVEEIVGDVRDEHDDATPDVVA
MAV_2878|M.avium_104 AEIRANSLQTALVVDEYGGTAGMVTVEDLIEEIVGDVRDEHDDATPDVVP
Mflv_3364|M.gilvum_PYR-GCK AEVRANGLQTALVVDEYGGTAGMVTVEDLIEEIVGDVRDEHDDEPPDVVA
Mvan_3094|M.vanbaalenii_PYR-1 SEVRANGLQTALVVDEYGGTAGMVTVEDLIEEIVGDVRDEHDVEPPDVVQ
TH_2608|M.thermoresistible__bu TQLRGNGLQTALVVDEYGGTAGMVTVEDMIEELVGDVRDEHDDATPAVVR
MSMEG_3637|M.smegmatis_MC2_155 TQIRANGLQTALVVDEYGGTAGMVTVEDLIEEIVGDVRDEHDDATPDVMP
MAB_2411c|M.abscessus_ATCC_199 EQVRANGMQTALVVDEYGGTAGLVTVEDMIEEIVGDVRDEHDDATPDVVS
MLBr_00387|M.leprae_Br4923 AGVGAPQITAILEAVAACGPAGVPVIADGGLQYSGDIAKALAAGASTTML
: . : : : . *.**: .: * : **: . .. .:
Mb1873c|M.bovis_AF2122/97 A---GNGWRVSGLLRIDEVASATGYR--APDGPYETIGG-----------
Rv1842c|M.tuberculosis_H37Rv A---GNGWRVSGLLRIDEVASATGYR--APDGPYETIGG-----------
MMAR_2716|M.marinum_M A---GNGWRVSGLLRIDEVAIAIGYQ--APEGPYETIGG-----------
MAV_2878|M.avium_104 A---GTGWRVSGLLRIDEVAAATGFR--APEGPYETIGG-----------
Mflv_3364|M.gilvum_PYR-GCK A---GQGWQVSGLLRIDEVAEGTAFR--APEGDYETIGG-----------
Mvan_3094|M.vanbaalenii_PYR-1 A---GRGWQVSGLLRIDEVAQGTEFR--APEGDYETIGG-----------
TH_2608|M.thermoresistible__bu A---PHGWQVSGLLRIDEVADQTGFR--PPEGDYETIGG-----------
MSMEG_3637|M.smegmatis_MC2_155 A---GKGWQVSGLLRIDEVAAQTGFR--APEGEYETIGG-----------
MAB_2411c|M.abscessus_ATCC_199 S---GGGWVVSGLLRIDEVAAATGYR--APEGEYETIGG-----------
MLBr_00387|M.leprae_Br4923 GSLLAGTAEAPGELIFVNGKQFKSYRGMGSLGAMQGRGGDKSYSKDRYFA
. ..* * : : :: . * : **
Mb1873c|M.bovis_AF2122/97 -LVLRELGHIPVAGETVELTALDQDGLPDDSMRWLATVIQMDGR-RIDSL
Rv1842c|M.tuberculosis_H37Rv -LVLRELGHIPVAGETVELTALDQDGLPDDSMRWLATVIQMDGR-RIDLL
MMAR_2716|M.marinum_M -LVLRQLGHIPVAGETVELPALDEDRVPDQSVRWRATVVQMDGR-RIDLL
MAV_2878|M.avium_104 -LVLREIGHIPAAGETVKLPALDPDGLLDPVLRWQARVIRMDGR-RIDVL
Mflv_3364|M.gilvum_PYR-GCK -LILEKLGHIPDEGETVELTAFDPDGAIEDPVRWLATVIKMDGR-RIDQL
Mvan_3094|M.vanbaalenii_PYR-1 -LVLEKLGHIPEEGESVELIAFDPDGPIQDPVHWLATVVKMDGR-RIDQL
TH_2608|M.thermoresistible__bu -LVLQELGHIPEAGEAVELTAFEPDGPMEDPVRWLATVVQMEGR-RIDLL
MSMEG_3637|M.smegmatis_MC2_155 -LVLQELGHIPETGEAVDLTAFDPDTDADEPTHWLATVVRMDGR-RIDLL
MAB_2411c|M.abscessus_ATCC_199 -LVLQELGHIPTVGETVELTAVVPDHPLAELPRWRARVAGMDGR-RIDQI
MLBr_00387|M.leprae_Br4923 DDALSEDKLVPEGIEGRVPFRGPLSSVIHQLVGGLRAAMGYTGSPTIEVL
* : :* * . . * *: :
Mb1873c|M.bovis_AF2122/97 ELIKMG----GHADPGSGRGR------------
Rv1842c|M.tuberculosis_H37Rv ELIKMG----GHADPGSGRGR------------
MMAR_2716|M.marinum_M ELTELAGAEDGQDDSDPERGGR-----------
MAV_2878|M.avium_104 ELTEID----AHAEAPGGRR-------------
Mflv_3364|M.gilvum_PYR-GCK RLTRLG----RLSDAPEGEDHRG----------
Mvan_3094|M.vanbaalenii_PYR-1 RLTELG----RKGDS------RG----------
TH_2608|M.thermoresistible__bu ELVELG----RGNGGPKAGVQR-----------
MSMEG_3637|M.smegmatis_MC2_155 ELTKLE----PAGEGEPNG--------------
MAB_2411c|M.abscessus_ATCC_199 ELTELT-----DADSRGNNDDG-----------
MLBr_00387|M.leprae_Br4923 QQAQFVRITPAGLKESHPHDVAMTVEAPNYYPR
. .: