For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVGYRTLDVPEGVHRGMPSSMLTFIVSLDDGVEAADTATALPAARPTPLILGGLHLRASHVRQRRGQAGV QLAVHPLASRALFGLPSAELPVAEYDATAVLGRDIVDLHHRVAEAHRWPDVFALVAQYLIDARRRRDGAT VRPEVLHAWHLLQRSRGLMPVTALADAVGVTTRHLATLFRREVGHSPKTVAMLMRFQHATGRIAESARRH GRVDLARVAADTGYSDQAHLSREFVRFAGVPPQRWLAEEFRNIQDGGHLLGSQWEHDCFESDRLVDTASP
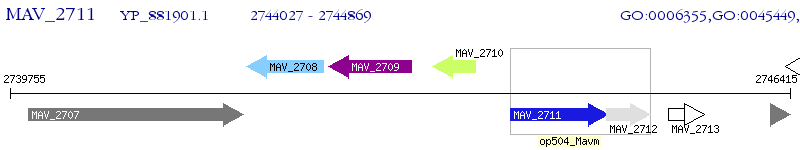
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2711 | - | - | 100% (280) | transcriptional regulator, AraC family protein |
| M. avium 104 | MAV_0221 | - | 1e-16 | 30.95% (252) | transcriptional regulator, AraC family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0534 | - | 1e-05 | 36.47% (85) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4712 | - | 1e-101 | 64.49% (276) | putative transcriptional regulator, AraC |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3998 | - | 6e-06 | 30.21% (96) | MmsAB operon regulatory protein |
| M. thermoresistible (build 8) | TH_2588 | - | 3e-31 | 34.66% (251) | PUTATIVE putative transcriptional regulator, AraC family |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3996 | - | 4e-05 | 29.27% (82) | helix-turn-helix domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2711|M.avium_104 -------------------------------------------MVGYRTL
MAB_4712|M.abscessus_ATCC_1997 -----------------MAGNEAPNWDFVVAPVRSVPGVAS--MVGYRAR
TH_2588|M.thermoresistible__bu ------------------VRDPARIEIGGHPAAPLRPFVTG--YTGFDLR
Mvan_3996|M.vanbaalenii_PYR-1 -------------------MDATGVDRRVPARLVLATRDGQ--LPGADLR
Mflv_0534|M.gilvum_PYR-GCK MTQDPESEARPSALVFDGATDSVGELRLPQRESELLPMANGPKMAGSYFS
MSMEG_3998|M.smegmatis_MC2_155 ---------------MDESTAAWIADGFVGQRMLAVPRPVVDHALGAPVT
*
MAV_2711|M.avium_104 DVPEGVHRG-MPSSMLTFIVSLDDGVEAADTATALPAARPTP--------
MAB_4712|M.abscessus_ATCC_1997 DIPDGIHLG-MPSATLTFIVSLDDGVESADSAAALDDARPNP--------
TH_2588|M.thermoresistible__bu GFEPGVHVG-MPSHTLTAVLTFGEPLEIDVPSGLSGTSGRYP--------
Mvan_3996|M.vanbaalenii_PYR-1 VYQFHSHQTQVPPLRDIAVIGWQSRAQIALRCGAIRSRAQMSPG------
Mflv_0534|M.gilvum_PYR-GCK RRETVCTEQLSAAEHLASEIFTPHRLRYSRRGQRLNARLSAARIGAVTVG
MSMEG_3998|M.smegmatis_MC2_155 RRLLVTDAGFFPRATRHGRSRSRGATEHVVMVCTAGAGWCRTEEG-----
. . .
MAV_2711|M.avium_104 ------------------------------LILGGLHLRASHVRQR----
MAB_4712|M.abscessus_ATCC_1997 ------------------------------LVLGGLHTRAPHIRQR----
TH_2588|M.thermoresistible__bu ------------------------------SMASGLLAQSISIRHR----
Mvan_3996|M.vanbaalenii_PYR-1 -----------------DFSILRPGYESTWQWSRGFQATVLYLNERRFAD
Mflv_0534|M.gilvum_PYR-GCK YLRYGADVELVNSTALDDYHINVPLTSHADSWCGPATAQATPTQAAVFLP
MSMEG_3998|M.smegmatis_MC2_155 ------------------------------AFDVGVGDAVLLPKGRAHMY
.
MAV_2711|M.avium_104 RGQAG--------VQLAVHPLASRALFGLPSAELPVAEYDATAVLG--RD
MAB_4712|M.abscessus_ATCC_1997 PGQAG--------IQLAVHPLASRALFGVPSAELPVTDFQGLEILG--GA
TH_2588|M.thermoresistible__bu GAQRG--------VKLSFTPLGARMVFGMPAGALANSVVPLSDVLA--DL
Mvan_3996|M.vanbaalenii_PYR-1 FASEV--------FDHHVDSVTIRESFQVQDSVIHHGLFLLATELR--RD
Mflv_0534|M.gilvum_PYR-GCK GRPAG--------IKWAADCGQLCVKFSRTDLEYELEGLLGRPAVRPLNI
MSMEG_3998|M.smegmatis_MC2_155 GAAESDPWTLWWMHVTGSDSAELIAAARVSPVSPVVHLVDPAPATSLISQ
MAV_2711|M.avium_104 IVDLHHRVAEAHRWPDVFALVAQYLIDARRRRDGA---TVRPEVLHAWHL
MAB_4712|M.abscessus_ATCC_1997 VAGLPERTARQRRWSDAFDTVATYLTESRRQRSVA---PVRSEVVQAWHL
TH_2588|M.thermoresistible__bu WPELEDRLGAAATWPHRFRVLDDVLARAVARTMCAGMFDVRPEVAAAWQR
Mvan_3996|M.vanbaalenii_PYR-1 EFGAALYRDALVTQLCVHLLRHHAESRLRAHRHPGALNAAQAQQVTDYIE
Mflv_0534|M.gilvum_PYR-GCK THSMDLTADSSRAWLALLSVLYREVSRPESIVQHPMTGRHFEQLVMHGFL
MSMEG_3998|M.smegmatis_MC2_155 VIDMLADASTTAGLIGAAGAAWHALTVIAATGRQRSTGEADPVDRALEHL
MAV_2711|M.avium_104 LQR-------------------------------SRGLMPVTALADAVGV
MAB_4712|M.abscessus_ATCC_1997 LER-------------------------------SQGRITVSTLASRVGI
TH_2588|M.thermoresistible__bu LVD-------------------------------SRGQVRVGDLAHDLGC
Mvan_3996|M.vanbaalenii_PYR-1 HN--------------------------------LDSDLCLSSLARVAGL
Mflv_0534|M.gilvum_PYR-GCK HAQPHYQAMALAADQHAAPPPKVLRTALDLIEEHPERSWTTTDLAREVGI
MSMEG_3998|M.smegmatis_MC2_155 RST-------------------------------SPRRTDVAALAAIVGL
** *
MAV_2711|M.avium_104 TTRHLATLFRREVGHSPKTVAMLMRFQHATGRIAESARRHGRVDLARVAA
MAB_4712|M.abscessus_ATCC_1997 SSRYLTTLFQREVGRSPKTVAMLMRFQRATSLLALSARR-GRVDLAAIAA
TH_2588|M.thermoresistible__bu SRRHLTARFREEFGLTPKTLARILRLEQAVGLVAAAGAP----AWAEVSA
Mvan_3996|M.vanbaalenii_PYR-1 SQYHFARLFKKRFGAPPHAYVQRRRIERARHLVLHSDMP-----LKEVVG
Mflv_0534|M.gilvum_PYR-GCK SARALQESFKRHVGLPPLTYLREVRLIRAHAELAASSES--AVTVARVAM
MSMEG_3998|M.smegmatis_MC2_155 STSHLNALFRQRTGIGPLEFQQQLRMSRARELLDTTTLT-----IAAVAR
: : *:.. * * *: :* : : :
MAV_2711|M.avium_104 DTGYSDQAHLSREFVRFAGVPPQRWLAEEFRNIQDGGHLLGSQWEHDCFE
MAB_4712|M.abscessus_ATCC_1997 ETGFSDQAHLTHEFVRFTGVPPRTWLAAEFRNIQDGGHSFGSECEHDYLD
TH_2588|M.thermoresistible__bu AAGYADQAHLVRDWRSFTGRTPSSWFDG---DVLVDGRPHG---------
Mvan_3996|M.vanbaalenii_PYR-1 VTGFCDQSHLTRAFRRAFDITPAELRRSSD--------------------
Mflv_0534|M.gilvum_PYR-GCK KWGFGHLGRFSAEYRRKFGVTPQHTLRAA---------------------
MSMEG_3998|M.smegmatis_MC2_155 AAGFNDPLYFSRQFAKVHGQSPSAYRQRHQN-------------------
*: . : : . .*
MAV_2711|M.avium_104 SDRLVDTASP
MAB_4712|M.abscessus_ATCC_1997 AQRVADP---
TH_2588|M.thermoresistible__bu ----------
Mvan_3996|M.vanbaalenii_PYR-1 ----------
Mflv_0534|M.gilvum_PYR-GCK ----------
MSMEG_3998|M.smegmatis_MC2_155 ----------