For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSCPILFAICENDTVAPAKATQKYAAQAPRGEIKLYDAGHFDIYVGADFERNVTDQLNFLSRHVPVS
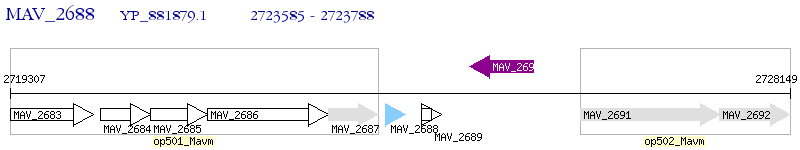
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2688 | - | - | 100% (67) | alpha/beta hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3976 | - | 6e-09 | 44.07% (59) | dienelactone hydrolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2608 | - | 2e-07 | 38.81% (67) | putative hydrolase, alpha/beta fold |
| M. marinum M | MMAR_0385 | - | 9e-19 | 59.38% (64) | hypothetical protein MMAR_0385 |
| M. smegmatis MC2 155 | MSMEG_4450 | - | 3e-11 | 42.19% (64) | alpha/beta hydrolase |
| M. thermoresistible (build 8) | TH_1408 | - | 5e-10 | 39.06% (64) | alpha/beta hydrolase |
| M. ulcerans Agy99 | MUL_1035 | - | 6e-17 | 54.41% (68) | hypothetical protein MUL_1035 |
| M. vanbaalenii PYR-1 | Mvan_2419 | - | 2e-08 | 42.62% (61) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0385|M.marinum_M -----MTQRQDVRFPCGEAMISAWLYRPAGADPARPVPMLVMAHGLGAVR
MUL_1035|M.ulcerans_Agy99 -----MTQRQDVRFPCGEALISAWLYRPAGADPARPVPMLVMAHGLGAVR
MAV_2688|M.avium_104 --------------------------------------------------
MSMEG_4450|M.smegmatis_MC2_155 MAQPLSYTRDDVTFMSDGTSCAAWLYRP---DGVNNPPIVVLAHGFAAFR
TH_1408|M.thermoresistible__bu MAEHT-YVRHDTTFISEGDRCAAWLYRP---DGVQNPPLVVMAHGFAAFR
Mflv_3976|M.gilvum_PYR-GCK -----MPTRDDVRIPAHGDEIAAYVYRPP--AGVGATACVVMAHGFTGTR
Mvan_2419|M.vanbaalenii_PYR-1 ------MTRQEVQIPADGGQIAAYVYRPQ--VTGGGTACIVMAHGFTGTR
MAB_2608|M.abscessus_ATCC_1997 ------MNRQSIRFNSHGIDCEAWLYRAQ--TVEDTAPIVVMAHGVGGTK
MMAR_0385|M.marinum_M TMRLDAYAQRFSAAGYACLVFDYRNFGDSEGQPRQLLDIRMQLQDWAAAV
MUL_1035|M.ulcerans_Agy99 TMRLDAYAQRFSAAGYACLVFDYRNFGDSEGQPRQLLDIRMQLQDWAAAV
MAV_2688|M.avium_104 --------------------------------------------------
MSMEG_4450|M.smegmatis_MC2_155 ELRLDAYAARFAQAGYAALVFDYRHWGASDGQPRRILDIPKQQTDWRAAI
TH_1408|M.thermoresistible__bu ELRLDAYADRFARAGYAVLVFDYRFWGASDGEPRRILDIKAQQADWRAAV
Mflv_3976|M.gilvum_PYR-GCK DDGLPDYAEAFRDAGYVVVLFDYRHFGASSGQPRQLLDMARQREDFHTVI
Mvan_2419|M.vanbaalenii_PYR-1 DDGLPDYAEAFCAAGHTVVLFDYRHFGASTGEPRQLLDIAEQRRDYHTVI
MAB_2608|M.abscessus_ATCC_1997 DSGLEPFAVRLAEAGMHALAFDYRGFGASDGFPRQHVSFTHQIGDYHAAV
MMAR_0385|M.marinum_M AYARTCDGVDQARIGLWGTSFGGGHVIATAARLPGIAAVVAQCPFTDGIA
MUL_1035|M.ulcerans_Agy99 AYARTCDGVDQARIGLWGTSFGGGHVIATAARLPGIAAVVAQCPFTDGIA
MAV_2688|M.avium_104 --------------------------------------------------
MSMEG_4450|M.smegmatis_MC2_155 AYARSLDNVDTTRLVGWGSSFGGGHVLTLAARDHEFAAAIVQVPHVSGPA
TH_1408|M.thermoresistible__bu SYARSLDSVDTTRLVAWGSSFGGGHVLHLLAHDHELAAGIAQVPHVDGPA
Mflv_3976|M.gilvum_PYR-GCK EWARRLDGVDPNRIVAWGSSFSGGHVLAVAAEDPRLAAVIAQAPFTDALA
Mvan_2419|M.vanbaalenii_PYR-1 AWARHLDGVDPNRIVVWGSSFSGGHVLAVAAEDPRIAAVIAQAPFTDALA
MAB_2608|M.abscessus_ATCC_1997 NMARRLPGVDPTRIVLWGVSLAGGHVMAVAGARNDVAAVIALTPLVDGIA
MMAR_0385|M.marinum_M SVLAIAPTTAIRVTARSMRDLIGSRLGRPPVMIPTAGKPGEVALMTAPDA
MUL_1035|M.ulcerans_Agy99 SVLAIAPTTAIRVTARSMRDLIGSRLGRPPVMIPTAGKPGEVALMTAPDA
MAV_2688|M.avium_104 --------------------------------------------------
MSMEG_4450|M.smegmatis_MC2_155 SAFSQSPKLVARLIAAGLRDQIGAWFGGAPHRVTSIGRPGEFAMMTSPGA
TH_1408|M.thermoresistible__bu SAFAQPIGLVSRLFLAGVRDQVGAWFGAEPHRTAAVGRPGELAMMTSPGA
Mflv_3976|M.gilvum_PYR-GCK TLRNAPLRNIPPMVVAALRDQLGAWRNRPPRMLAAVGPPGDLAALTSPDA
Mvan_2419|M.vanbaalenii_PYR-1 TLREIPPGNIPLLLIAAVRDQVGAWRGRPPHTVAAVGSPGTVAALTSPDA
MAB_2608|M.abscessus_ATCC_1997 AARSAMTEHSAGTLLRSAVTGLRSRLGRQQRTIPIVGRAGELAALTSDGY
MMAR_0385|M.marinum_M YPGYLGLMP--------QGTQVPNEVAARIGIKVLGYRPGRSARKIACPI
MUL_1035|M.ulcerans_Agy99 YPGYLGLMP--------QGTQVPNEVAARIGIKVLGYRPGRSARKIACPI
MAV_2688|M.avium_104 ---------------------------------------------MSCPI
MSMEG_4450|M.smegmatis_MC2_155 YELVEEMAGGEAAEHMRAQLEAENDVAARIALRVPLYSPGRHAAKITAPT
TH_1408|M.thermoresistible__bu MDLVEQMAA-----DHREKLFTENNVAARIALRVPFYSPGRYASRITAPT
Mflv_3976|M.gilvum_PYR-GCK EPGFGAILG--------PESLWRNEFAARLMLRFAFERPGRKAATLQMPV
Mvan_2419|M.vanbaalenii_PYR-1 KPGFEAIIG--------PNSLWRNEFAARLMFTFAFNRPGRKAAKLRMPV
MAB_2608|M.abscessus_ATCC_1997 YEAHLAIAG----------PAWRNEIDATVGTQLVSYRPTRCASRIGCPI
: *
MMAR_0385|M.marinum_M LFCVCETDSVAPAGPTLRYAARAPRG----EVIRYPEGHFAIYLGDAF-E
MUL_1035|M.ulcerans_Agy99 LFCVCETDSVAPAGPTLRYAARAPRGEVSGEVIRYPEGHFAIYLGDAF-E
MAV_2688|M.avium_104 LFAICENDTVAPAKATQKYAAQAPRG----EIKLYDAGHFDIYVGADF-E
MSMEG_4450|M.smegmatis_MC2_155 LVQLATKDDVTPYAKARKIVARIPNG----EVRSYDITHFEPYVDPHF-E
TH_1408|M.thermoresistible__bu LIQVARYDDVTPMDKALRVARRIPRG----EALVYDCRHFEPYLEPHF-E
Mflv_3976|M.gilvum_PYR-GCK LFCVCDDDVVTPPGPTIEAARRAPCG----ELRRYPYGHFDIYHDP----
Mvan_2419|M.vanbaalenii_PYR-1 LLCVCDDDVVTPPGPSVEAAHRAPRG----ELCRYPYGHFDIYHDP----
MAB_2608|M.abscessus_ATCC_1997 LVQIADFDRAAPPHAAAKAAFKARAE-----VRHYPCDHFGVYHGQECHE
*. :. * .:* : . . : * ** *
MMAR_0385|M.marinum_M RVVADQLGFLDKHLKPAGQP-------
MUL_1035|M.ulcerans_Agy99 HVVADQLGFLDKHLKPAGQP-------
MAV_2688|M.avium_104 RNVTDQLNFLSRHVPVS----------
MSMEG_4450|M.smegmatis_MC2_155 QIVTDQIDFLNRHVGAGTR--------
TH_1408|M.thermoresistible__bu KIVSDQIDFLHRHVGQR----------
Mflv_3976|M.gilvum_PYR-GCK QVKADQLEFLHRALARGGR--------
Mvan_2419|M.vanbaalenii_PYR-1 KVKADQLEFLARVVGDRTA--------
MAB_2608|M.abscessus_ATCC_1997 LVIGHQISFLSRHLSIEAVSHTEALHG
.*: ** : :