For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVNRALQGLDLTTLDLVIIENVGNLVCPAEFDVGEHAKAMVYSVTKGDDKPMKYPVMFRAVDVVLLNKID LVPYLDVDVDSYIGHVREVNPTATILPVSARTGVGMPDWLAWLRRFMEGTPTA
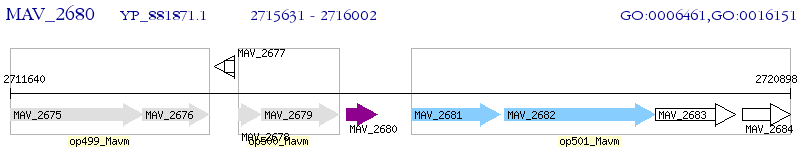
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2680 | hypB | - | 100% (123) | hydrogenase accessory protein HypB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3978 | - | 2e-30 | 53.10% (113) | hydrogenase accessory protein HypB |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1871 | hypB | 7e-55 | 80.99% (121) | nickel cation-binding GTPase, HypB |
| M. smegmatis MC2 155 | MSMEG_2271 | hypB | 2e-51 | 82.05% (117) | hydrogenase accessory protein HypB |
| M. thermoresistible (build 8) | TH_2818 | - | 3e-54 | 82.35% (119) | PUTATIVE nickel cation-binding GTPase, HypB |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2357 | - | 3e-49 | 75.86% (116) | hydrogenase accessory protein HypB |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1871|M.marinum_M -MGRFHRHDDGTVHTHEH-----EH-HQHG---DHTGYQT--GPARIEVL
TH_2818|M.thermoresistible__bu -VGRFHRHDDGTVHSHEHGDHAHEHPHDHG---DHSGYQT--GGQRIEVL
Mvan_2357|M.vanbaalenii_PYR-1 -MGRFHRHDDGTAHTHDHDG---SPSHDHG---DHSGYHT--AAERVDVL
MSMEG_2271|M.smegmatis_MC2_155 -MGRFHRHDDGTVHTHDHDHDGHVHDHAHG---DHSGYQT--GTERIEVL
MAV_2680|M.avium_104 --------------------------------------------------
Mflv_3978|M.gilvum_PYR-GCK MCATCGCGEDGTTITVPVQGHDHHHNHPHDQPDDHHQHHHPHPTETVTLE
MMAR_1871|M.marinum_M EEIFAENDVRADLNRRAFETNGIRALNLMSSPGSGKTTVLQATLDQLASE
TH_2818|M.thermoresistible__bu ESIFAENDSRAAINRKIFEENGIRALNLMSSPGSGKTTVLAATLDELANQ
Mvan_2357|M.vanbaalenii_PYR-1 EAIFSENDLRAAANRNAFEENGIRALNLMSSPGSGKTSLLAVTLDELVGE
MSMEG_2271|M.smegmatis_MC2_155 ESIFAENDIRADINRGLFAEHGVRALNLMSSPGSGKTTLLARTLDDLGAD
MAV_2680|M.avium_104 --------------------------------------------------
Mflv_3978|M.gilvum_PYR-GCK QKVLAKNDGIAARNRRWLTERDIVAFNMTSSPGSGKTTLLERTIRDLHPT
MMAR_1871|M.marinum_M FAIGVIEGDIATDLDAAKLSGRGAQVSLLNTSNGFGGECHLDAPMVNRAL
TH_2818|M.thermoresistible__bu IAIGIIEGDIATDLDAAKLGGRGAQISLLNTNNGFGGECHLDAPMVNRAL
Mvan_2357|M.vanbaalenii_PYR-1 IAIGVIEGDIATDLDAARLEGRGAQISLLNTDNGFGGECHLDAPMVHRAL
MSMEG_2271|M.smegmatis_MC2_155 VSVGIVEGDIETDLDAAKLGGRGAQISLLNTANGFGGECHLDAPMVNRAL
MAV_2680|M.avium_104 --------------------------------------------MVNRAL
Mflv_3978|M.gilvum_PYR-GCK RSVAVIEGDQETLLDAERIQATGARVVQVNTGAG----CHLDAEMIRRAL
*:.***
MMAR_1871|M.marinum_M PGLDLPNLDLVIVENVGNLVCPAEFDVGEHAKAMVYSVTEGEDKPLKYPV
TH_2818|M.thermoresistible__bu QDLDLDHLELVVIENVGNLVCPAEFDVGEHAKAMVYSVAEGEDKPLKYPV
Mvan_2357|M.vanbaalenii_PYR-1 AGLNLAGLDLVVIENVGNLVCPAEFDVGEHAKVMVYSLTEGEDKPLKYPV
MSMEG_2271|M.smegmatis_MC2_155 QGLDLTRLDLVIIENVGNLVCPAEFDVGEHAKVMVYSVTEGEDKPLKYPV
MAV_2680|M.avium_104 QGLDLTTLDLVIIENVGNLVCPAEFDVGEHAKAMVYSVTKGDDKPMKYPV
Mflv_3978|M.gilvum_PYR-GCK DTLAPRRARVLFIENVGNLVCPALFDLGETSKVVVISVTEGADKPLKYPH
* ::.:********** **:** :*.:* *:::* ***:***
MMAR_1871|M.marinum_M MFRCVDVVLLNKIDLVPYLDADVDTYVAHIREVNAHATILPVSARTGAGM
TH_2818|M.thermoresistible__bu MFRAVDVVLLNKIDLVPYLDVDVDRYIAHIREVNPTARILPVSARTGEGM
Mvan_2357|M.vanbaalenii_PYR-1 MFRAVDVVLLNKIDLAPYLDADVATYTERIRQVNPTATVLPVSAKTGEGM
MSMEG_2271|M.smegmatis_MC2_155 MFRAVDVVLLNKIDLVAHLDADVRTYTSRVREVNPTTEILPVSARTGEGM
MAV_2680|M.avium_104 MFRAVDVVLLNKIDLVPYLDVDVDSYIGHVREVNPTATILPVSARTGVGM
Mflv_3978|M.gilvum_PYR-GCK MFAAADLVLINKSDLLPYVDFDIDACASAARSVNPGVGILTLSAKTGDGV
** ..*:**:** ** .::* *: *.**. . :*.:**:** *:
MMAR_1871|M.marinum_M TAWFDWLRR---------FAQGLPPE-----------
TH_2818|M.thermoresistible__bu DAWFDWLRQ---------FLRG---------------
Mvan_2357|M.vanbaalenii_PYR-1 SEWYRWLRA---------FAKD---------------
MSMEG_2271|M.smegmatis_MC2_155 EAWYDWLRR---------FLTAGR-------------
MAV_2680|M.avium_104 PDWLAWLRR---------FMEGTPTA-----------
Mflv_3978|M.gilvum_PYR-GCK EEWYDWISQRSIYDARVPIAGGRPAPEVRPHQEIRRR
* *: :