For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSGAVRAAAIAGVALSDVGRVDDKGPYELIAQASRRALADAGLTPADIDGLASTGQGTLPPTDVGEYLGL RPRWIDSTAVGGASWEVMAAHAADAIAAGHADVVLLTYGSTARSDLRKGLRGANINWGARGPLQWEAPYG HTLISKYAMAARRHMFTYGTTIEQLAEVAVSARFNAAENPEAYYRDPIAIDDVLSGPMIADPFTKLHCCI RSDGGAAVVLVSAERAKDLRAKPVWVLGSGETTSHMLTSQWDDLTVGPASVSGPLAFARAGVSPSDVDVA ELYDAFTYMLLLTLEDLGFCPKGEGGPFVEQGALRLGGELPTNTDGGGLSACHPGQRGLFLLVEAARQLR GECGPRQVPDARIACVSGTGGWFCSSGTMILGTEEP
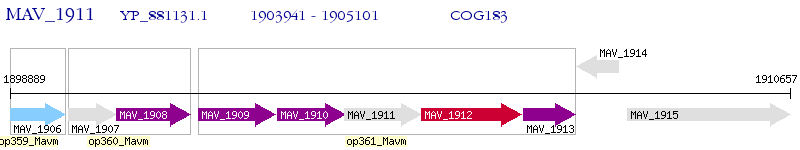
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1911 | - | - | 100% (386) | thiolase |
| M. avium 104 | MAV_2621 | - | 0.0 | 93.78% (386) | thiolase |
| M. avium 104 | MAV_0477 | - | 9e-60 | 38.83% (394) | lipid-transfer protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3570c | ltp2 | 1e-48 | 35.03% (374) | lipid-transfer protein |
| M. gilvum PYR-GCK | Mflv_2483 | - | 9e-55 | 38.85% (399) | hypothetical protein Mflv_2483 |
| M. tuberculosis H37Rv | Rv3540c | ltp2 | 1e-48 | 35.03% (374) | lipid-transfer protein |
| M. leprae Br4923 | MLBr_01158 | fadA4 | 8e-06 | 24.84% (161) | acetyl-CoA acetyltransferase |
| M. abscessus ATCC 19977 | MAB_0618 | - | 4e-57 | 37.80% (381) | lipid-transfer protein |
| M. marinum M | MMAR_5027 | ltp2_1 | 4e-50 | 35.47% (375) | acetyl-CoA acetyltransferase (PaaJ-like), Ltp2_1 |
| M. smegmatis MC2 155 | MSMEG_5990 | - | 2e-53 | 35.53% (380) | lipid-transfer protein |
| M. thermoresistible (build 8) | TH_4525 | - | 1e-132 | 60.89% (381) | PUTATIVE thiolase |
| M. ulcerans Agy99 | MUL_4101 | ltp2_1 | 4e-51 | 35.73% (375) | lipid-transfer protein |
| M. vanbaalenii PYR-1 | Mvan_0899 | - | 6e-56 | 38.82% (389) | lipid-transfer protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1911|M.avium_104 --------------------------------------------------
TH_4525|M.thermoresistible__bu --------------------------------------------------
MMAR_5027|M.marinum_M --------------------------------------------------
MUL_4101|M.ulcerans_Agy99 --------------------------------------------------
Mb3570c|M.bovis_AF2122/97 --------------------------------------------------
Rv3540c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0618|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_5990|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0899|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2483|M.gilvum_PYR-GCK MSRLPLPLLTFDNEFFWTSGADGVLRIQECGDCKSLIHPPQPVCRYCHSH
MLBr_01158|M.leprae_Br4923 --------------------------------------------------
MAV_1911|M.avium_104 --------------------------------------------------
TH_4525|M.thermoresistible__bu --------------------------------------------------
MMAR_5027|M.marinum_M --------------------------------------------------
MUL_4101|M.ulcerans_Agy99 --------------------------------------------------
Mb3570c|M.bovis_AF2122/97 --------------------------------------------------
Rv3540c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0618|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_5990|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0899|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2483|M.gilvum_PYR-GCK NMGPREVSGRAVLSAFTVNHRFSIPGLEAPYVVAQVAIEEDPRVRLTTNI
MLBr_01158|M.leprae_Br4923 --------------------------------------------------
MAV_1911|M.avium_104 --------------------------------------------------
TH_4525|M.thermoresistible__bu --------------------------------------------------
MMAR_5027|M.marinum_M --------------------------------------------------
MUL_4101|M.ulcerans_Agy99 --------------------------------------------------
Mb3570c|M.bovis_AF2122/97 --------------------------------------------------
Rv3540c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0618|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_5990|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0899|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2483|M.gilvum_PYR-GCK VDCDPDELELGRVVEVVFEQNEDVWLPLFRPTAEPETAALPDDEIAPQEF
MLBr_01158|M.leprae_Br4923 --------------------------------------------------
MAV_1911|M.avium_104 ---------MSGAVRAAAIAG--VALSDVGRVDDKGPYELIAQASRRALA
TH_4525|M.thermoresistible__bu -----------MAAKGVAIVG--VGLSDCGRVPDKTATALMAQAARRAIS
MMAR_5027|M.marinum_M -----------MLSGKAAIVG--IGATDFSKNSGRSELRLAAEAVRDALD
MUL_4101|M.ulcerans_Agy99 -----------MLSGKAAIVG--IGATDFSKNSGRSELRLAAEAVRDALD
Mb3570c|M.bovis_AF2122/97 -----------MLSGQAAIVG--IGATDFSKNSGRSELRLAAEAVLDALA
Rv3540c|M.tuberculosis_H37Rv -----------MLSGQAAIVG--IGATDFSKNSGRSELRLAAEAVLDALA
MAB_0618|M.abscessus_ATCC_1997 ---------MTGLSGRSAIAG--IGATDFSKKSGRSELRLAAEAVLDALD
MSMEG_5990|M.smegmatis_MC2_155 ---------MTGLSGKAAIAG--IGATDFSKNSGRSELRLAAECVLDALD
Mvan_0899|M.vanbaalenii_PYR-1 -----------MTQDRCAVVG--VGNTDYSFNSGRSTLTLAAQAGVAALN
Mflv_2483|M.gilvum_PYR-GCK PKFVRPMLTTEKFEDAAAITG--IGASKMGRRLMVSPLSLTVEACEKAIA
MLBr_01158|M.leprae_Br4923 ------MTTSVIVTGARTPIGKLMGSLKDFSASDLGAITIAAALKKANVA
: * :. . : . :
MAV_1911|M.avium_104 DAGLTPADIDGLASTGQGTLPP------TDVGEYLGLRPRWIDSTAVGGA
TH_4525|M.thermoresistible__bu DAGLHKNDIDGLGAHGS-LLAP------IEVGEYLGLRPTWVDSTDVGGS
MMAR_5027|M.marinum_M DAGLSPADVDGLTTFTMDTNT----EIAVARAAGIGELKFFSK-IHYGGG
MUL_4101|M.ulcerans_Agy99 DAGLSPADVDGLTTFTMDTNT----EIAVARAAGIGELKFFSK-IHYGGG
Mb3570c|M.bovis_AF2122/97 DAGLSPTDVDGLTTFTMDTNT----EIAVARAAGIGELTFFSK-IHYGGG
Rv3540c|M.tuberculosis_H37Rv DAGLSPTDVDGLTTFTMDTNT----EIAVARAAGIGELTFFSK-IHYGGG
MAB_0618|M.abscessus_ATCC_1997 DAGLSPSDVDGLVTFTMDSNL----ETAVARATGIGELKFFSQ-IGYGGG
MSMEG_5990|M.smegmatis_MC2_155 DAGLEPSDVDGLVTFTMDSNQ----ETAVARSTGIGELKFFSQ-IGYGGG
Mvan_0899|M.vanbaalenii_PYR-1 DAGLEPADIDGIVRCEMDEVL----HADLAATLGMTNVNFWAA-GSWAGA
Mflv_2483|M.gilvum_PYR-GCK DAGLTLADIDGLSTYPAMDAMGMGEGGCTVLEGALGIRPTWIN-GGMDTF
MLBr_01158|M.leprae_Br4923 PSIVQYVIIGQVLTAGAGQMPAR--QAAVAAGIGWDVPALTINKMCLSGL
: : :. :
MAV_1911|M.avium_104 SWEVMAAHAADA------IAAGHADVVLLTYGST-ARSDLRKGLRGANIN
TH_4525|M.thermoresistible__bu SWEVMAQHAVAA------IAAGEIDVALLTYGST-ARSDVKRRKRAPAAA
MMAR_5027|M.marinum_M AACATVQHAAMA------VATGIADVVVAYRAFN-ERSGMRFGQVQTRLT
MUL_4101|M.ulcerans_Agy99 AACATVQHAAMA------VATGIADVVVAYRAFN-ERSDMRFGQVQTRLT
Mb3570c|M.bovis_AF2122/97 AACATVQHAAMA------VATGVADVVVAYRAFN-ERSGMRFGQVQTRLT
Rv3540c|M.tuberculosis_H37Rv AACATVQHAAMA------VATGVADVVVAYRAFN-ERSGMRFGQVQTRLT
MAB_0618|M.abscessus_ATCC_1997 AAAATVQQAALA------VAAGVAEVVVAYRAFN-ERSEFRFGQVMTGLS
MSMEG_5990|M.smegmatis_MC2_155 AAAATVQQAALA------VATGVAEVVVAYRAFN-ERSEFRFGQVMTGLT
Mvan_0899|M.vanbaalenii_PYR-1 APCGMVGLAVNA------ICSGQATNVLVFRSLN-GRSGRRFGREWS-SG
Mflv_2483|M.gilvum_PYR-GCK GPGGSLIAAVMA------VATGMARHVLCFRTLW-EAT---FNQLMKEGK
MLBr_01158|M.leprae_Br4923 DAIALADQLIRAGEFDVVVAGGQESMTKAPHLLMDSRSGYKYGDVTIVDH
* :. * . :
MAV_1911|M.avium_104 WGARG---PLQWEAPYGHT-LISKYAMAARRHMFTYGTTIEQLAEVAVSA
TH_4525|M.thermoresistible__bu MPTGG---AMQYETPYGAT-LIAKYAMAARRHMIEYGTTIDQLAEVAVAA
MMAR_5027|M.marinum_M ENADSTGVDNSFSYPHGLSTPAAQVAMIAKRYMHLSGASSRDFGAVSVAD
MUL_4101|M.ulcerans_Agy99 ENADSTGVDNSFSYPHGLSTPAAQVAMIAKRYMHLSGASSRDFGAVSVAD
Mb3570c|M.bovis_AF2122/97 ENADSTGVDNSFSYPHGLSTPAAQVAMIARRYMHLSGATSRDFGAVSVAD
Rv3540c|M.tuberculosis_H37Rv ENADSTGVDNSFSYPHGLSTPAAQVAMIARRYMHLSGATSRDFGAVSVAD
MAB_0618|M.abscessus_ATCC_1997 STADSRGVEYSWSYPHGLSTPAASVAMIARRYMHEYGATSADFGAVSVAD
MSMEG_5990|M.smegmatis_MC2_155 TNADSRGVEYSWSYPHGLSTPAASVAMIARRYMHEYGATSADFGVVSVAD
Mvan_0899|M.vanbaalenii_PYR-1 QTVGGAGSYLEFLAPYGLQSPGMAYALFARKHMIDYGTTSEQLGHIAVTC
Mflv_2483|M.gilvum_PYR-GCK ASPPGGARVNSWQAPFGATSAAHTLALNAQRHFHRYGTTKETLGWIALNQ
MLBr_01158|M.leprae_Br4923 LAYDGLHDVFTNQPMGALTEQRNDVEKFTRQEQDEFAARSHQKAAAAWKD
. . ::: .: . :
MAV_1911|M.avium_104 RFNAAENP-EAYYRD-----PIAIDDVLSGPMIADPFTKLHCCIRSDGGA
TH_4525|M.thermoresistible__bu HEWAAMNE-NAFDRE-----RLTIEDVAAAPMLADPFTTRHVCLRTDGGG
MMAR_5027|M.marinum_M RKHAAKNPKAYFYQK-----PITIDDHQNSRWIAEPLRLLDCCQETDGAV
MUL_4101|M.ulcerans_Agy99 RKHAAKNPKAYFYQK-----PITIDDHQNSRWIAEPLRLLDCCQETDGAV
Mb3570c|M.bovis_AF2122/97 RKHAANNPKAYFYGK-----PITIEDHQNSRWIAEPLRLLDCCQETDGAV
Rv3540c|M.tuberculosis_H37Rv RKHAANNPKAYFYGK-----PITIEDHQNSRWIAEPLRLLDCCQETDGAV
MAB_0618|M.abscessus_ATCC_1997 RKHAATNPKAFFYGK-----PITIEDHQNSRMIADPVRLLDCCQESDGGV
MSMEG_5990|M.smegmatis_MC2_155 RKHAANNPKAHFYGK-----PITIEDHQNSRWIAEPLRLLDCCQETDGGV
Mvan_0899|M.vanbaalenii_PYR-1 HRRANQNPTAQRYGH-----PLTMAGYLESRMIADPLRRFDFCLETDGAA
Mflv_2483|M.gilvum_PYR-GCK RANAALNP-TAIYRD-----PMTMQDYLNARPITTPFGLYDCDVPCDGAV
MLBr_01158|M.leprae_Br4923 GVFADEVVPVSIPQSKGDSLQFTEDEGIRANTSAESLAGLKPAFRCGG--
* :: . : .. . .*
MAV_1911|M.avium_104 AVVLVSAERAKDLRAKPVWVLGSGETTS----HMLTSQWDDLTVGPASVS
TH_4525|M.thermoresistible__bu AVVLAGERIARDCAGEPVWILGTASAAS----HVSMDEWADFTTSAAAES
MMAR_5027|M.marinum_M AIVVTSAERARDLQQRPAIIEAASQGSS----PDQYTMVSYYRPELDGLP
MUL_4101|M.ulcerans_Agy99 AIVVTSAERARDLQQRPAIIEAASQGSS----PDQYTMVSYYRPELDGLP
Mb3570c|M.bovis_AF2122/97 AIVVTSAARARDLKQRPVVIEAAAQGCS----PDQYTMVSYYRPELDGLP
Rv3540c|M.tuberculosis_H37Rv AIVVTSAARARDLKQRPVVIEAAAQGCS----PDQYTMVSYYRPELDGLP
MAB_0618|M.abscessus_ATCC_1997 AIVVTTPERAKDLKNRPVIIEAAAQGSG----EDQFTMYSYYRDELG-LP
MSMEG_5990|M.smegmatis_MC2_155 AIVVTTPERAKDLKHRPVTIEAAAQGSG----ADQFTMYSYYRDELG-LP
Mvan_0899|M.vanbaalenii_PYR-1 AVIVSSAERARDGAATPILIRAVVQGDE----PVPRQGGLWGNLLTGPLT
Mflv_2483|M.gilvum_PYR-GCK AVIVSAVDAAADAPKPPVFFEAVGTQII----ERTDWDQTTLTHEPQVLG
MLBr_01158|M.leprae_Br4923 TITPGSASQISDGAATVVVMNKEKAQQLGLTWLVEIGAHGVVAGPDSTLQ
:: * .
MAV_1911|M.avium_104 GPL------AFARAGVSPSDVDVAELYDAFTYMLLLTLEDLGFCPKGEGG
TH_4525|M.thermoresistible__bu GPK------AFARAGVTPADIDVCQLYDSFTSTVLLTVEDLGFCAKGEGG
MMAR_5027|M.marinum_M EMG-LVGQQLWQQSGLKPTDIQTAVLYDHFTPFTLIQLEELGFCGKGEAK
MUL_4101|M.ulcerans_Agy99 EMG-LVGQQLWQQSGLKPTDIQTAVLYDHFTPFTLIQLEELGFCGKGEAK
Mb3570c|M.bovis_AF2122/97 EMG-LVGRQLWAQSGLTPADVQTAVLYDHFTPFTLIQLEELGFCGKGEAK
Rv3540c|M.tuberculosis_H37Rv EMG-LVGRQLWAQSGLTPADVQTAVLYDHFTPFTLIQLEELGFCGKGEAK
MAB_0618|M.abscessus_ATCC_1997 EMG-LVGRQLWDQSGLTPNDIQTAILYDHFTPYTLLQLEELGFCGKGEAK
MSMEG_5990|M.smegmatis_MC2_155 EMG-LVGRQLWQQSGLTPDDIQTAILYDHFTPYTLLQLEELGFCGKGEAK
Mvan_0899|M.vanbaalenii_PYR-1 NASSALARRLYQTAGMGPDDIDVAQFYDCFTITVLMQLEDYGFCARGEGG
Mflv_2483|M.gilvum_PYR-GCK QAA-----HLWTRTSLRPDDVDVAELYDGFTFNCLSWLEALGFCGIGESK
MLBr_01158|M.leprae_Br4923 SQPANAIKKAVDREGISVEQLDVVEINEAFAAVALASARELGIAPELVNV
.: :::. : : *: * . *:.
MAV_1911|M.avium_104 PFVEQG-ALRLGGELPTNTDGGGLSACHPGQRGLFLLVEAARQLRGECGP
TH_4525|M.thermoresistible__bu PFIASG-TLRPDGALPTNTDGGGLAACHPGMRGMFLMVEAVRQLRGECGP
MMAR_5027|M.marinum_M DFIADG-AIELGGRLPINTHGGQLGEAY--IHGMNGIAEGVRQLRGTS-V
MUL_4101|M.ulcerans_Agy99 DFIADG-AIELGGRLPINTHGGQLGEAY--IHGMNGIAEGVRQLRGTS-V
Mb3570c|M.bovis_AF2122/97 DFIADG-AIEVGGRLPINTHGGQLGEAY--IHGMNGIAEGVRQLRGTS-V
Rv3540c|M.tuberculosis_H37Rv DFIADG-AIEVGGRLPINTHGGQLGEAY--IHGMNGIAEGVRQLRGTS-V
MAB_0618|M.abscessus_ATCC_1997 DFIANG-AIELGGKLPINTHGGQLGEAY--IHGMNGIAEGVRQLRGTS-V
MSMEG_5990|M.smegmatis_MC2_155 DFIAGG-AIEIGGKLPINTHGGQLGEAY--IHGMNGIAEGVRQLRGTS-V
Mvan_0899|M.vanbaalenii_PYR-1 PFAASG-ALELDGALPINTSGGNLADGY--IHGLSHIVEGVRQLRGQS-T
Mflv_2483|M.gilvum_PYR-GCK DFLDGGHAIARDGVIPLNTHGGQLSHGR--THGMGLVHEAVSQLRGEAGE
MLBr_01158|M.leprae_Br4923 N----------GGAIAVGHPLG--------MSGARITLHVALELARRG--
.* :. . * * . . :*
MAV_1911|M.avium_104 RQVPDARIACVSGTGGWFCSSGTMILGTEEP
TH_4525|M.thermoresistible__bu RQVADAELACVHAMGGFFSHSATMILGRDR-
MMAR_5027|M.marinum_M NPVPDVEHVLVT-AGTGVP-TSGLILG----
MUL_4101|M.ulcerans_Agy99 NPVPDVEHVLVT-AGTGVP-TSGLILG----
Mb3570c|M.bovis_AF2122/97 NPVAGVEHVLVT-AGTGVP-TSGLILG----
Rv3540c|M.tuberculosis_H37Rv NPVAGVEHVLVT-AGTGVP-TSGLILG----
MAB_0618|M.abscessus_ATCC_1997 NQVPNVEHVLVT-AGTGVP-TSGLILG----
MSMEG_5990|M.smegmatis_MC2_155 NQVEGVEHVLVT-AGTGVP-TSGLILG----
Mvan_0899|M.vanbaalenii_PYR-1 ANVPDARTCLVT-SGMPGPGSSALILRTD--
Mflv_2483|M.gilvum_PYR-GCK RQVADARVAVVS-SGGLTP-SGVMLLRTGA-
MLBr_01158|M.leprae_Br4923 ---SGYGVAALCGAGGQGD---ALILRAV--
. : * ::*