For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ISLRNQAAIVGIGHVPFGRRGELAEKGHLRLAVEAITLACEDAGITGKDVDGYSSYYTEVEPAELVSAFG AQRLRYTAQTWGGGGASMCGAFQNAALGVTTGQADYVVVHKVATMAGTARYGQAFGQMGQGVNAIGGPFA FSVPYGLQSPGQMFALAARRHMHLYGTTSEQFAHVAINARTMAANNPAARFREPITVADHQASPMIADPL RRLDFCMESDYGCAVVIASADRARDLRQKPAYIAAAAVGAPYRYGNALMSFYNQPDEDFASSGHRTVADE LYRHSGLTLQDMHAAMLYDHFTAMMIMALEDFGFCKKGEGGPYVADGNLALGGPLPVNTHGGNLAEAYTH GMTHVMEAVRQIRGSAINQIPSATNILVVGGAGPSPTSGMILTGSR*
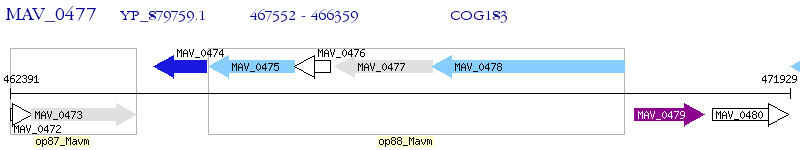
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0477 | - | - | 100% (397) | lipid-transfer protein |
| M. avium 104 | MAV_1918 | - | 2e-85 | 44.92% (394) | thiolase |
| M. avium 104 | MAV_2630 | - | 8e-85 | 44.90% (392) | thiolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3570c | ltp2 | 4e-81 | 43.26% (393) | lipid-transfer protein |
| M. gilvum PYR-GCK | Mflv_1513 | - | 2e-79 | 41.84% (392) | lipid-transfer protein |
| M. tuberculosis H37Rv | Rv3540c | ltp2 | 4e-81 | 43.26% (393) | lipid-transfer protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0618 | - | 2e-79 | 41.27% (395) | lipid-transfer protein |
| M. marinum M | MMAR_5027 | ltp2_1 | 2e-81 | 42.49% (393) | acetyl-CoA acetyltransferase (PaaJ-like), Ltp2_1 |
| M. smegmatis MC2 155 | MSMEG_5990 | - | 5e-77 | 40.56% (392) | lipid-transfer protein |
| M. thermoresistible (build 8) | TH_4616 | - | 5e-89 | 46.19% (394) | PUTATIVE putative nonspecific lipid-transfer protein |
| M. ulcerans Agy99 | MUL_4101 | ltp2_1 | 1e-81 | 42.49% (393) | lipid-transfer protein |
| M. vanbaalenii PYR-1 | Mvan_5253 | - | 2e-78 | 41.58% (392) | lipid-transfer protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0618|M.abscessus_ATCC_1997 --MTGLSGRSAIAGIGATDFSKKSG---RSELRLAAEAVLDALDDAGLSP
MSMEG_5990|M.smegmatis_MC2_155 --MTGLSGKAAIAGIGATDFSKNSG---RSELRLAAECVLDALDDAGLEP
Mflv_1513|M.gilvum_PYR-GCK ----MLSGKAAIAGIGATDFSKNSG---RSELRLAAEAVLDALDDAGLTT
Mvan_5253|M.vanbaalenii_PYR-1 ----MLSGKAAIAGIGATDFSKNSG---RSELRLAAEAVLDALDDAGLTP
Mb3570c|M.bovis_AF2122/97 ----MLSGQAAIVGIGATDFSKNSG---RSELRLAAEAVLDALADAGLSP
Rv3540c|M.tuberculosis_H37Rv ----MLSGQAAIVGIGATDFSKNSG---RSELRLAAEAVLDALADAGLSP
MMAR_5027|M.marinum_M ----MLSGKAAIVGIGATDFSKNSG---RSELRLAAEAVRDALDDAGLSP
MUL_4101|M.ulcerans_Agy99 ----MLSGKAAIVGIGATDFSKNSG---RSELRLAAEAVRDALDDAGLSP
MAV_0477|M.avium_104 --MISLRNQAAIVGIGHVPFGRRGELAEKGHLRLAVEAITLACEDAGITG
TH_4616|M.thermoresistible__bu VDSNGLR-DVAIVGVGATPFYKRGGSLPKTIVELAGEAIIAACEDAGLPV
* **.*:* . * :.. : :.** *.: * ***:
MAB_0618|M.abscessus_ATCC_1997 SDVDGLVTFT------MDSNLETAVARATGIGELKFFSQIGYGGGAA-AA
MSMEG_5990|M.smegmatis_MC2_155 SDVDGLVTFT------MDSNQETAVARSTGIGELKFFSQIGYGGGAA-AA
Mflv_1513|M.gilvum_PYR-GCK ADVDGLVTFS------MDSNLETAVARSTGIGELKFFSQIGYGGGAA-AA
Mvan_5253|M.vanbaalenii_PYR-1 SDVDGLVTFT------MDSNLETAVARSTGIGDLTFFSQIGYGGGAA-AA
Mb3570c|M.bovis_AF2122/97 TDVDGLTTFT------MDTNTEIAVARAAGIGELTFFSKIHYGGGAA-CA
Rv3540c|M.tuberculosis_H37Rv TDVDGLTTFT------MDTNTEIAVARAAGIGELTFFSKIHYGGGAA-CA
MMAR_5027|M.marinum_M ADVDGLTTFT------MDTNTEIAVARAAGIGELKFFSKIHYGGGAA-CA
MUL_4101|M.ulcerans_Agy99 ADVDGLTTFT------MDTNTEIAVARAAGIGELKFFSKIHYGGGAA-CA
MAV_0477|M.avium_104 KDVDGYSSY------YT-EVEPAELVSAFGAQRLRYTAQTWGGGGASMCG
TH_4616|M.thermoresistible__bu HDIDGFAYYSGANAGYTDKMDTADFMETLGIPEVRFTAALTSGGGGS-AG
*:** : . : * : : : ***.: ..
MAB_0618|M.abscessus_ATCC_1997 TVQQAALAVAAGVAEVVVAYRAFNERSEFRFGQVMTGLSSTADSRGVEYS
MSMEG_5990|M.smegmatis_MC2_155 TVQQAALAVATGVAEVVVAYRAFNERSEFRFGQVMTGLTTNADSRGVEYS
Mflv_1513|M.gilvum_PYR-GCK TVQQAALAVATGVAEVVVAYRAFNERSEHRFGQVMTGLTVNADSRGVEYS
Mvan_5253|M.vanbaalenii_PYR-1 TVQQAALAVATGVAEVVVAYRAFNERSEHRFGQVMTGLTVNADSRGVEYS
Mb3570c|M.bovis_AF2122/97 TVQHAAMAVATGVADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNS
Rv3540c|M.tuberculosis_H37Rv TVQHAAMAVATGVADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNS
MMAR_5027|M.marinum_M TVQHAAMAVATGIADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNS
MUL_4101|M.ulcerans_Agy99 TVQHAAMAVATGIADVVVAYRAFNERSDMRFGQVQTRLTENADSTGVDNS
MAV_0477|M.avium_104 AFQNAALGVTTGQADYVVVHKVATMAGTARYGQAFGQMGQGVNAIGGPFA
TH_4616|M.thermoresistible__bu AIGLARAAIVAGDAS-VVVTVMALQQAKQRLGSVFSAMER--DPIN---S
:. * .:.:* *. **. . * *.. : :. . :
MAB_0618|M.abscessus_ATCC_1997 WSYPHGLSTPAASVAMIARRYMHEYGATSADFGAVSVADRKHAATNPKAF
MSMEG_5990|M.smegmatis_MC2_155 WSYPHGLSTPAASVAMIARRYMHEYGATSADFGVVSVADRKHAANNPKAH
Mflv_1513|M.gilvum_PYR-GCK WSYPHGLSTPAASVAMIAQRYMHEFGATSADFGVISVADRKHAANNPKAH
Mvan_5253|M.vanbaalenii_PYR-1 WSYPHGLSTPAASVAMIARRYMHEYGATSADFGAVSVADRKHAATNPKAH
Mb3570c|M.bovis_AF2122/97 FSYPHGLSTPAAQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAY
Rv3540c|M.tuberculosis_H37Rv FSYPHGLSTPAAQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAY
MMAR_5027|M.marinum_M FSYPHGLSTPAAQVAMIAKRYMHLSGASSRDFGAVSVADRKHAAKNPKAY
MUL_4101|M.ulcerans_Agy99 FSYPHGLSTPAAQVAMIAKRYMHLSGASSRDFGAVSVADRKHAAKNPKAY
MAV_0477|M.avium_104 FSVPYGLQSPGQMFALAARRHMHLYGTTSEQFAHVAINARTMAANNPAAR
TH_4616|M.thermoresistible__bu FLQPSGLFGPGQLMSVLARRHMHLYGTRREAFAEIAISTRANALNRPNAM
: * ** *. .:: *:*:** *: *. ::: * * ..* *
MAB_0618|M.abscessus_ATCC_1997 FYGKPITIEDHQNSRMIADPVRLLDCCQESDGGVAIVVTTPERAKDLKNR
MSMEG_5990|M.smegmatis_MC2_155 FYGKPITIEDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERAKDLKHR
Mflv_1513|M.gilvum_PYR-GCK FYGKPITIEDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERARDLKHR
Mvan_5253|M.vanbaalenii_PYR-1 FYGKPITIEDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERAKDLRHR
Mb3570c|M.bovis_AF2122/97 FYGKPITIEDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAARARDLKQR
Rv3540c|M.tuberculosis_H37Rv FYGKPITIEDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAARARDLKQR
MMAR_5027|M.marinum_M FYQKPITIDDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAERARDLQQR
MUL_4101|M.ulcerans_Agy99 FYQKPITIDDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAERARDLQQR
MAV_0477|M.avium_104 FR-EPITVADHQASPMIADPLRRLDFCMESDYGCAVVIASADRARDLRQK
TH_4616|M.thermoresistible__bu YR-EPLTLENYFNARMIAEPLCLYDFCLETDGAVAVITTSMDRARDLRQP
: :*:*: :: : **:*: * * *:* . *:: :: **:**::
MAB_0618|M.abscessus_ATCC_1997 PVIIEAAAQGSGEDQF-TMYSYYRDELG-LPEMG--LVGRQLWDQSGLTP
MSMEG_5990|M.smegmatis_MC2_155 PVTIEAAAQGSGADQF-TMYSYYRDELG-LPEMG--LVGRQLWQQSGLTP
Mflv_1513|M.gilvum_PYR-GCK PAIIEAAAQGAGADQF-TMYSYYREELG-LPEMG--LVGRQLWAQSGLTP
Mvan_5253|M.vanbaalenii_PYR-1 PAVIEAAAQGAGADQF-TMYSYYRDELG-LPEMG--LVGRQLWEQSGLSP
Mb3570c|M.bovis_AF2122/97 PVVIEAAAQGCSPDQY-TMVSYYRPELDGLPEMG--LVGRQLWAQSGLTP
Rv3540c|M.tuberculosis_H37Rv PVVIEAAAQGCSPDQY-TMVSYYRPELDGLPEMG--LVGRQLWAQSGLTP
MMAR_5027|M.marinum_M PAIIEAASQGSSPDQY-TMVSYYRPELDGLPEMG--LVGQQLWQQSGLKP
MUL_4101|M.ulcerans_Agy99 PAIIEAASQGSSPDQY-TMVSYYRPELDGLPEMG--LVGQQLWQQSGLKP
MAV_0477|M.avium_104 PAYIAAAAVGAPYRYGNALMSFYNQPDEDFASSGHRTVADELYRHSGLTL
TH_4616|M.thermoresistible__bu PVPVIAAAHGGVRDWG-RAFAWMGMADDYFASSGNRPIADRLYRQADITA
*. : **: * :: :.. * :. .*: ::.:.
MAB_0618|M.abscessus_ATCC_1997 NDIQTAILYDHFTPYTLLQLEELGFCGKGEAKDFIANGAIELGG-KLPIN
MSMEG_5990|M.smegmatis_MC2_155 DDIQTAILYDHFTPYTLLQLEELGFCGKGEAKDFIAGGAIEIGG-KLPIN
Mflv_1513|M.gilvum_PYR-GCK QDIQTAVLYDHFTPYTLIQLEELGFCGKGEAKDFIAGGAIEIGG-RLPIN
Mvan_5253|M.vanbaalenii_PYR-1 EDIQTAILYDHFTPYTLIQLEELGFCGKGEAKDFIAGGAIEIGG-RLPIN
Mb3570c|M.bovis_AF2122/97 ADVQTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIEVGG-RLPIN
Rv3540c|M.tuberculosis_H37Rv ADVQTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIEVGG-RLPIN
MMAR_5027|M.marinum_M TDIQTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIELGG-RLPIN
MUL_4101|M.ulcerans_Agy99 TDIQTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIELGG-RLPIN
MAV_0477|M.avium_104 QDMHAAMLYDHFTAMMIMALEDFGFCKKGEGGPYVADGNLALGG-PLPVN
TH_4616|M.thermoresistible__bu DDIDVALLYDHFTPMVLMQLEDYGFCGKGEGGAFVESGAIRYDGGAIPVN
*:..*:******. :: **: *** ***. :: .* : .* :*:*
MAB_0618|M.abscessus_ATCC_1997 THGGQLGEAYIHGMNGIAEGVRQLRGTSVNQVPNVEHVLVTAGTG-VPTS
MSMEG_5990|M.smegmatis_MC2_155 THGGQLGEAYIHGMNGIAEGVRQLRGTSVNQVEGVEHVLVTAGTG-VPTS
Mflv_1513|M.gilvum_PYR-GCK THGGQLGEAYIHGMNGIAEGVRQLRGTSVNQVPDVEHVLVTAGTG-VPTS
Mvan_5253|M.vanbaalenii_PYR-1 THGGQLGEAYIHGMNGIAEGVRQLRGTSVNQVPDVEHVLVTAGTG-VPTS
Mb3570c|M.bovis_AF2122/97 THGGQLGEAYIHGMNGIAEGVRQLRGTSVNPVAGVEHVLVTAGTG-VPTS
Rv3540c|M.tuberculosis_H37Rv THGGQLGEAYIHGMNGIAEGVRQLRGTSVNPVAGVEHVLVTAGTG-VPTS
MMAR_5027|M.marinum_M THGGQLGEAYIHGMNGIAEGVRQLRGTSVNPVPDVEHVLVTAGTG-VPTS
MUL_4101|M.ulcerans_Agy99 THGGQLGEAYIHGMNGIAEGVRQLRGTSVNPVPDVEHVLVTAGTG-VPTS
MAV_0477|M.avium_104 THGGNLAEAYTHGMTHVMEAVRQIRGSAINQIPSATNILVVGGAGPSPTS
TH_4616|M.thermoresistible__bu PHGGQLSEAYIVGMTHIMEGVEQMRGTAINQVPNAELALVTGGPASLPVS
.***:*.*** **. : *.*.*:**:::* : .. **..*.. *.*
MAB_0618|M.abscessus_ATCC_1997 GLILG---
MSMEG_5990|M.smegmatis_MC2_155 GLILG---
Mflv_1513|M.gilvum_PYR-GCK GLILG---
Mvan_5253|M.vanbaalenii_PYR-1 GLILG---
Mb3570c|M.bovis_AF2122/97 GLILG---
Rv3540c|M.tuberculosis_H37Rv GLILG---
MMAR_5027|M.marinum_M GLILG---
MUL_4101|M.ulcerans_Agy99 GLILG---
MAV_0477|M.avium_104 GMILTGSR
TH_4616|M.thermoresistible__bu GLILGKAA
*:**