For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSGQAAIVGIGATDFSKNSGRSELRLAAEAVLDALADAGLSPTDVDGLTTFTMDTNTEIAVARAAGIGEL TFFSKIHYGGGAACATVQHAAMAVATGVADVVVAYRAFNERSGMRFGQVQTRLTENADSTGVDNSFSYPH GLSTPAAQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAYFYGKPITIEDHQNSRWIAEPLRLLD CCQETDGAVAIVVTSAARARDLKQRPVVIEAAAQGCSPDQYTMVSYYRPELDGLPEMGLVGRQLWAQSGL TPADVQTAVLYDHFTPFTLIQLEELGFCGKGEAKDFIADGAIEVGGRLPINTHGGQLGEAYIHGMNGIAE GVRQLRGTSVNPVAGVEHVLVTAGTGVPTSGLILG*
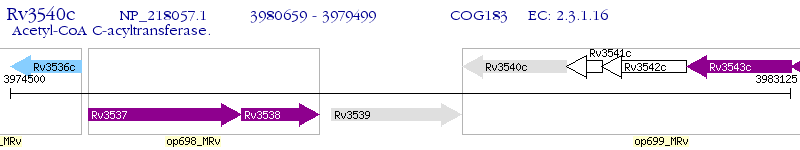
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3540c | ltp2 | - | 100% (386) | lipid-transfer protein |
| M. tuberculosis H37Rv | Rv2790c | ltp1 | 1e-26 | 26.82% (399) | lipid-transfer protein |
| M. tuberculosis H37Rv | Rv1627c | - | 2e-24 | 33.73% (252) | lipid-transfer protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3570c | ltp2 | 0.0 | 100.00% (386) | lipid-transfer protein |
| M. gilvum PYR-GCK | Mflv_1513 | - | 0.0 | 84.46% (386) | lipid-transfer protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0618 | - | 0.0 | 82.86% (385) | lipid-transfer protein |
| M. marinum M | MMAR_5027 | ltp2_1 | 0.0 | 93.26% (386) | acetyl-CoA acetyltransferase (PaaJ-like), Ltp2_1 |
| M. avium 104 | MAV_0621 | - | 0.0 | 89.49% (390) | lipid-transfer protein |
| M. smegmatis MC2 155 | MSMEG_5990 | - | 0.0 | 84.68% (385) | lipid-transfer protein |
| M. thermoresistible (build 8) | TH_0366 | ltp2 | 0.0 | 91.71% (386) | PROBABLE LIPID TRANSFER PROTEIN OR KETO ACYL-COA THIOLASE |
| M. ulcerans Agy99 | MUL_4101 | ltp2_1 | 0.0 | 93.01% (386) | lipid-transfer protein |
| M. vanbaalenii PYR-1 | Mvan_5253 | - | 0.0 | 84.46% (386) | lipid-transfer protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0618|M.abscessus_ATCC_1997 MTGLSGRSAIAGIGATDFSKKSGRSELRLAAEAVLDALDDAGLSPSDVDG
MSMEG_5990|M.smegmatis_MC2_155 MTGLSGKAAIAGIGATDFSKNSGRSELRLAAECVLDALDDAGLEPSDVDG
Mvan_5253|M.vanbaalenii_PYR-1 --MLSGKAAIAGIGATDFSKNSGRSELRLAAEAVLDALDDAGLTPSDVDG
Mflv_1513|M.gilvum_PYR-GCK --MLSGKAAIAGIGATDFSKNSGRSELRLAAEAVLDALDDAGLTTADVDG
Rv3540c|M.tuberculosis_H37Rv --MLSGQAAIVGIGATDFSKNSGRSELRLAAEAVLDALADAGLSPTDVDG
Mb3570c|M.bovis_AF2122/97 --MLSGQAAIVGIGATDFSKNSGRSELRLAAEAVLDALADAGLSPTDVDG
MMAR_5027|M.marinum_M --MLSGKAAIVGIGATDFSKNSGRSELRLAAEAVRDALDDAGLSPADVDG
MUL_4101|M.ulcerans_Agy99 --MLSGKAAIVGIGATDFSKNSGRSELRLAAEAVRDALDDAGLSPADVDG
MAV_0621|M.avium_104 --MLSRKAAIVGIGATDFSKDSGRSELRLAAEAVLDALDDAGLSPADVDG
TH_0366|M.thermoresistible__bu --MLSGKAAIVGIGATDFSKDSGRSELRLAAEAVLDALNDAGLTPADVDG
** ::**.*********.***********.* *** **** .:****
MAB_0618|M.abscessus_ATCC_1997 LVTFTMDSNLETAVARATGIGELKFFSQIGYGGGAAAATVQQAALAVAAG
MSMEG_5990|M.smegmatis_MC2_155 LVTFTMDSNQETAVARSTGIGELKFFSQIGYGGGAAAATVQQAALAVATG
Mvan_5253|M.vanbaalenii_PYR-1 LVTFTMDSNLETAVARSTGIGDLTFFSQIGYGGGAAAATVQQAALAVATG
Mflv_1513|M.gilvum_PYR-GCK LVTFSMDSNLETAVARSTGIGELKFFSQIGYGGGAAAATVQQAALAVATG
Rv3540c|M.tuberculosis_H37Rv LTTFTMDTNTEIAVARAAGIGELTFFSKIHYGGGAACATVQHAAMAVATG
Mb3570c|M.bovis_AF2122/97 LTTFTMDTNTEIAVARAAGIGELTFFSKIHYGGGAACATVQHAAMAVATG
MMAR_5027|M.marinum_M LTTFTMDTNTEIAVARAAGIGELKFFSKIHYGGGAACATVQHAAMAVATG
MUL_4101|M.ulcerans_Agy99 LTTFTMDTNTEIAVARAAGIGELKFFSKIHYGGGAACATVQHAAMAVATG
MAV_0621|M.avium_104 LTTFTMDTNNETAVARAVGIGDLKFFSQIGYGGGAACATVQQAAIAVATG
TH_0366|M.thermoresistible__bu LTTFTMDTNTEVAVARAVGIPELKFFSKIHYGGGAACATVQQAAMAVATG
*.**:**:* * ****:.** :*.***:* ******.****:**:***:*
MAB_0618|M.abscessus_ATCC_1997 VAEVVVAYRAFNERSEFRFGQVMTGLS----STADSRGVEYSWSYPHGLS
MSMEG_5990|M.smegmatis_MC2_155 VAEVVVAYRAFNERSEFRFGQVMTGLT----TNADSRGVEYSWSYPHGLS
Mvan_5253|M.vanbaalenii_PYR-1 VAEVVVAYRAFNERSEHRFGQVMTGLT----VNADSRGVEYSWSYPHGLS
Mflv_1513|M.gilvum_PYR-GCK VAEVVVAYRAFNERSEHRFGQVMTGLT----VNADSRGVEYSWSYPHGLS
Rv3540c|M.tuberculosis_H37Rv VADVVVAYRAFNERSGMRFGQVQTRLT----ENADSTGVDNSFSYPHGLS
Mb3570c|M.bovis_AF2122/97 VADVVVAYRAFNERSGMRFGQVQTRLT----ENADSTGVDNSFSYPHGLS
MMAR_5027|M.marinum_M IADVVVAYRAFNERSGMRFGQVQTRLT----ENADSTGVDNSFSYPHGLS
MUL_4101|M.ulcerans_Agy99 IADVVVAYRAFNERSDMRFGQVQTRLT----ENADSTGVDNSFSYPHGLS
MAV_0621|M.avium_104 VADVVVAYRAFNERSGMRFGQVQTRLAGDARAQADSTAADNSFSYPHGLS
TH_0366|M.thermoresistible__bu VADVVVAYRAFNERSGMRFGQVQTRLI----TGADSTSVDNSFSYPHGLS
:*:************ ***** * * *** ..: *:*******
MAB_0618|M.abscessus_ATCC_1997 TPAASVAMIARRYMHEYGATSADFGAVSVADRKHAATNPKAFFYGKPITI
MSMEG_5990|M.smegmatis_MC2_155 TPAASVAMIARRYMHEYGATSADFGVVSVADRKHAANNPKAHFYGKPITI
Mvan_5253|M.vanbaalenii_PYR-1 TPAASVAMIARRYMHEYGATSADFGAVSVADRKHAATNPKAHFYGKPITI
Mflv_1513|M.gilvum_PYR-GCK TPAASVAMIAQRYMHEFGATSADFGVISVADRKHAANNPKAHFYGKPITI
Rv3540c|M.tuberculosis_H37Rv TPAAQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAYFYGKPITI
Mb3570c|M.bovis_AF2122/97 TPAAQVAMIARRYMHLSGATSRDFGAVSVADRKHAANNPKAYFYGKPITI
MMAR_5027|M.marinum_M TPAAQVAMIAKRYMHLSGASSRDFGAVSVADRKHAAKNPKAYFYQKPITI
MUL_4101|M.ulcerans_Agy99 TPAAQVAMIAKRYMHLSGASSRDFGAVSVADRKHAAKNPKAYFYQKPITI
MAV_0621|M.avium_104 TPAAQVAMIARRYMHLSGATSRDFGAISVADRKHAAKNPKAYFYEKPITI
TH_0366|M.thermoresistible__bu TPAAQVAMIARRYMHLSGATSRDFGVISVADRKHAAKNPKAYFYQKPITI
****.*****:**** **:* ***.:*********.****.** *****
MAB_0618|M.abscessus_ATCC_1997 EDHQNSRMIADPVRLLDCCQESDGGVAIVVTTPERAKDLKNRPVIIEAAA
MSMEG_5990|M.smegmatis_MC2_155 EDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERAKDLKHRPVTIEAAA
Mvan_5253|M.vanbaalenii_PYR-1 EDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERAKDLRHRPAVIEAAA
Mflv_1513|M.gilvum_PYR-GCK EDHQNSRWIAEPLRLLDCCQETDGGVAIVVTTPERARDLKHRPAIIEAAA
Rv3540c|M.tuberculosis_H37Rv EDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAARARDLKQRPVVIEAAA
Mb3570c|M.bovis_AF2122/97 EDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAARARDLKQRPVVIEAAA
MMAR_5027|M.marinum_M DDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAERARDLQQRPAIIEAAS
MUL_4101|M.ulcerans_Agy99 DDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSAERARDLQQRPAIIEAAS
MAV_0621|M.avium_104 EDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSVERARDLKQRPAVIEAAA
TH_0366|M.thermoresistible__bu EDHQNSRWIAEPLRLLDCCQETDGAVAIVVTSVERARDLKHRPVVVEAAA
:****** **:*:********:**.******: **:**::**. :***:
MAB_0618|M.abscessus_ATCC_1997 QGSGEDQFTMYSYYRDELG-LPEMGLVGRQLWDQSGLTPNDIQTAILYDH
MSMEG_5990|M.smegmatis_MC2_155 QGSGADQFTMYSYYRDELG-LPEMGLVGRQLWQQSGLTPDDIQTAILYDH
Mvan_5253|M.vanbaalenii_PYR-1 QGAGADQFTMYSYYRDELG-LPEMGLVGRQLWEQSGLSPEDIQTAILYDH
Mflv_1513|M.gilvum_PYR-GCK QGAGADQFTMYSYYREELG-LPEMGLVGRQLWAQSGLTPQDIQTAVLYDH
Rv3540c|M.tuberculosis_H37Rv QGCSPDQYTMVSYYRPELDGLPEMGLVGRQLWAQSGLTPADVQTAVLYDH
Mb3570c|M.bovis_AF2122/97 QGCSPDQYTMVSYYRPELDGLPEMGLVGRQLWAQSGLTPADVQTAVLYDH
MMAR_5027|M.marinum_M QGSSPDQYTMVSYYRPELDGLPEMGLVGQQLWQQSGLKPTDIQTAVLYDH
MUL_4101|M.ulcerans_Agy99 QGSSPDQYTMVSYYRPELDGLPEMGLVGQQLWQQSGLKPTDIQTAVLYDH
MAV_0621|M.avium_104 QGSSPDQYTMVSYYRSELG-LPEMGVVGRQLWQQSGLKPTDIQTAVLYDH
TH_0366|M.thermoresistible__bu QGSSADQYTMTSYYRPELDGLPEMGLVGRQLWAQSGLTPDDIQTAVLYDH
**.. **:** **** **. *****:**:*** ****.* *:***:****
MAB_0618|M.abscessus_ATCC_1997 FTPYTLLQLEELGFCGKGEAKDFIANGAIELGGKLPINTHGGQLGEAYIH
MSMEG_5990|M.smegmatis_MC2_155 FTPYTLLQLEELGFCGKGEAKDFIAGGAIEIGGKLPINTHGGQLGEAYIH
Mvan_5253|M.vanbaalenii_PYR-1 FTPYTLIQLEELGFCGKGEAKDFIAGGAIEIGGRLPINTHGGQLGEAYIH
Mflv_1513|M.gilvum_PYR-GCK FTPYTLIQLEELGFCGKGEAKDFIAGGAIEIGGRLPINTHGGQLGEAYIH
Rv3540c|M.tuberculosis_H37Rv FTPFTLIQLEELGFCGKGEAKDFIADGAIEVGGRLPINTHGGQLGEAYIH
Mb3570c|M.bovis_AF2122/97 FTPFTLIQLEELGFCGKGEAKDFIADGAIEVGGRLPINTHGGQLGEAYIH
MMAR_5027|M.marinum_M FTPFTLIQLEELGFCGKGEAKDFIADGAIELGGRLPINTHGGQLGEAYIH
MUL_4101|M.ulcerans_Agy99 FTPFTLIQLEELGFCGKGEAKDFIADGAIELGGRLPINTHGGQLGEAYIH
MAV_0621|M.avium_104 FTPFTLIQLEELGFCGKGEAKDFIADGAIEIGGRLPINTHGGQLGEAYIH
TH_0366|M.thermoresistible__bu FTPFTLIQLEELGFCGKGEAKDFIADGAIEIGGRLPINTHGGQLGEAYIH
***:**:******************.****:**:****************
MAB_0618|M.abscessus_ATCC_1997 GMNGIAEGVRQLRGTSVNQVPNVEHVLVTAGTGVPTSGLILG
MSMEG_5990|M.smegmatis_MC2_155 GMNGIAEGVRQLRGTSVNQVEGVEHVLVTAGTGVPTSGLILG
Mvan_5253|M.vanbaalenii_PYR-1 GMNGIAEGVRQLRGTSVNQVPDVEHVLVTAGTGVPTSGLILG
Mflv_1513|M.gilvum_PYR-GCK GMNGIAEGVRQLRGTSVNQVPDVEHVLVTAGTGVPTSGLILG
Rv3540c|M.tuberculosis_H37Rv GMNGIAEGVRQLRGTSVNPVAGVEHVLVTAGTGVPTSGLILG
Mb3570c|M.bovis_AF2122/97 GMNGIAEGVRQLRGTSVNPVAGVEHVLVTAGTGVPTSGLILG
MMAR_5027|M.marinum_M GMNGIAEGVRQLRGTSVNPVPDVEHVLVTAGTGVPTSGLILG
MUL_4101|M.ulcerans_Agy99 GMNGIAEGVRQLRGTSVNPVPDVEHVLVTAGTGVPTSGLILG
MAV_0621|M.avium_104 GMNGIAEGVRQLRGTSVNPVPDVEHVLVTAGTGVPTSGLILG
TH_0366|M.thermoresistible__bu GMNGIAEGVRQLRGTSVNQVDNVEHVLVTAGTGVPTSGLILG
****************** * .********************