For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PEASTPTIDRLVRSRAAEFGGKPMVIDPGYRITYDQLDTATRELAAVFVQAGVGKGTRVGLIMPNNTRWV LIAIALTRIGAVLVPLSTLLRAGELVAQLRVAAVQFLVSVDEFRGHRYLDDVAAVRSELPALQQVWPTEQ LDAAAAGARAGQIVDAMTQTVTPADPLVIMFTSGSSGTPKGVWHSHGSALGAVQSGLAARCIDADSRLYL PMPFFWVGGFGSGILSALLAGATLVTEEIPRPETTLRLLESERVTLFRGWPDQAETLARHAGTVGADLSA LRPGSLQALLPPEQRARPGARATLFGMTEAFGPYSGYPADTDMPVSARGSCGKPFDGMEVRIVDPDTGAP VGAGTAGIIQIRGPHTLRGMCGRSREELFTVDGFYPTGDLGHLDDAGFLFYHGRADDMFKVSGATVYPSE VERALRTIDGVDGAVVTNVPGATGDRVGAAVVCRELTAAQLRAAARNLLSSFKVPSVWLVLRSDDDLPRG VTGKVDVRRLRELLADADRRQETRVQG*
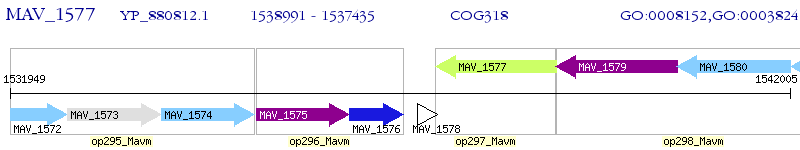
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1577 | - | - | 100% (518) | AMP-binding enzyme, putative |
| M. avium 104 | MAV_1579 | - | 2e-87 | 39.15% (516) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_0600 | fadD | 7e-50 | 32.13% (526) | acyl-CoA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3591 | fadD3 | 6e-52 | 33.46% (517) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_4364 | - | 0.0 | 66.40% (503) | AMP-dependent synthetase and ligase |
| M. tuberculosis H37Rv | Rv3561 | fadD3 | 1e-51 | 33.46% (517) | acyl-CoA synthetase |
| M. leprae Br4923 | MLBr_02546 | fadD2 | 2e-28 | 27.31% (498) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_4377c | - | 7e-44 | 29.53% (491) | AMP-binding domain protein |
| M. marinum M | MMAR_4027 | - | 0.0 | 71.97% (503) | fatty-acid-CoA ligase |
| M. smegmatis MC2 155 | MSMEG_2215 | - | 1e-177 | 64.29% (504) | AMP-binding enzyme |
| M. thermoresistible (build 8) | TH_4352 | - | 1e-128 | 76.92% (299) | PUTATIVE AMP-dependent synthetase and ligase |
| M. ulcerans Agy99 | MUL_4126 | fadD3 | 4e-52 | 32.82% (524) | acyl-CoA synthetase |
| M. vanbaalenii PYR-1 | Mvan_1980 | - | 0.0 | 65.27% (501) | AMP-dependent synthetase and ligase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3591|M.bovis_AF2122/97 ---------------------------------------MIND-LRTVPA
Rv3561|M.tuberculosis_H37Rv ---------------------------------------MIND-LRTVPA
MUL_4126|M.ulcerans_Agy99 ---------------------------------------MTSD-PRTVPT
MAV_1577|M.avium_104 ---------------------------------------MPEASTPTIDR
MMAR_4027|M.marinum_M ---------------------------------------MPD----TIDH
TH_4352|M.thermoresistible__bu --------------------------------------------------
Mflv_4364|M.gilvum_PYR-GCK ---------------------------------------MTD----TVDA
Mvan_1980|M.vanbaalenii_PYR-1 ---------------------------------------MTD----TIDS
MSMEG_2215|M.smegmatis_MC2_155 ---------------------------------------MTEH--LTVGA
MAB_4377c|M.abscessus_ATCC_199 ------------------------------MKAYDAGATLSPIIEETIGE
MLBr_02546|M.leprae_Br4923 MTKLRHYLERGAAEVHYLRSIFESGALGFEPPLTYAAMAVAMRKWGDLAM
Mb3591|M.bovis_AF2122/97 ALDRLVRQLPDHTALIAE--DRRFTSTELRDAVYGAAAALIALGVEPADR
Rv3561|M.tuberculosis_H37Rv ALDRLVRQLPDHTALIAE--DRRFTSTELRDAVYGAAAALIALGVEPADR
MUL_4126|M.ulcerans_Agy99 ALDRLVRRLPNHDALVTD--ERSFTVSELNDEVYRAAAALIELGVKPAAR
MAV_1577|M.avium_104 LVRSRAAEFGGKPMVIDP--GYRITYDQLDTATRELAAVFVQAGVGKGTR
MMAR_4027|M.marinum_M LVRSQAARHGAKPMVIDQ--ASRLSYRELDSSTSDLAAAFIQAGAGKGTR
TH_4352|M.thermoresistible__bu --------------------------------------------------
Mflv_4364|M.gilvum_PYR-GCK LLRAQAERHPDTEAVVDP--AERITYRELDAATRELGAAFVASGITKGTR
Mvan_1980|M.vanbaalenii_PYR-1 LLRDQAAAHGARDAVVDP--AERISYAELNRSTGELGAAFVASGIGKGMR
MSMEG_2215|M.smegmatis_MC2_155 LIRHHAADTPDKPMVVDP--KERIAYGDLDEKTRAMASGLVAAGVGKGSR
MAB_4377c|M.abscessus_ATCC_199 NFERIARAHPDVGALVDVSGDRRWTYRELDAEINRVARGLMSLGVAAGDR
MLBr_02546|M.leprae_Br4923 LPAMPARRSPHRAALIDE--EGTLTYKELDRAAHALANGLIAKGVRGGDG
Mb3591|M.bovis_AF2122/97 VAIWSPNTWHWVVACLAIHHAGAAVVPLNTRYTATEATDILDRAGAPVLF
Rv3561|M.tuberculosis_H37Rv VAIWSPNTWHWVVACLAIHHAGAAVVPLNTRYTATEATDILDRAGAPVLF
MUL_4126|M.ulcerans_Agy99 VGIWSPNTWHWVVACLAIHHVGAAMVPLNTRYTADEAADILARTEAPVLF
MAV_1577|M.avium_104 VGLIMPNNTRWVLIAIALTRIGAVLVPLSTLLRAGELVAQLRVAAVQFLV
MMAR_4027|M.marinum_M VGLIMPNGVRWVQIAIALTRIGAVLVPLSTLLRPRELASQLRSAAVQFLI
TH_4352|M.thermoresistible__bu --------------------------------------------------
Mflv_4364|M.gilvum_PYR-GCK VGLLMPNGVEWARTALALMRIGAVLVPLSTLLTPPELDAQLRTASVQHLI
Mvan_1980|M.vanbaalenii_PYR-1 VALLMPNGVAWARIALALMRFGAVVVPLSTLLTARELSAQLQTASVQHLI
MSMEG_2215|M.smegmatis_MC2_155 VGLLMPNGTDWVLTALAVTRIGAILVPLSTMLTPTELEAQLRTASVQFLI
MAB_4377c|M.abscessus_ATCC_199 VGIWAPNCAEWVLVQYATAKIGAILVNINPAYRTHELAYALNQSGVRTLI
MLBr_02546|M.leprae_Br4923 IAILARNHRWFVIANYGCARVGARIILLNSEFSGPQIKEVSEREGAKVII
Mb3591|M.bovis_AF2122/97 AAGLFLGADRAAGLDRA---ALPALRHVVRVPVEADDGTWDEFIATGAGA
Rv3561|M.tuberculosis_H37Rv AAGLFLGADRAAGLDRA---ALPALRHVVRVPVEADDGTWDEFIATGAGA
MUL_4126|M.ulcerans_Agy99 AMGHFLGSDRVAGLDRK---ALPALRHIVRIPIEEPDGTWDEFIETGT-D
MAV_1577|M.avium_104 SVDEFRGHRYLDDVAAV-RSELPALQQVWPTEQLDA-------AAAGARA
MMAR_4027|M.marinum_M SVPEFRGHRYLDAVESVPRAELPALQRVWADDQVSS-------SNAGARA
TH_4352|M.thermoresistible__bu --------------------------------------------------
Mflv_4364|M.gilvum_PYR-GCK AVEEFRGHRYLDRLDAD----LPALSTVWTTDQITG-------LRPGPGT
Mvan_1980|M.vanbaalenii_PYR-1 AVEEFRGHRY--HLDAT----PPALRSVRTPEQVFA-------LRGHPAA
MSMEG_2215|M.smegmatis_MC2_155 AVDEFRGRGYPVAPERL-----PALRQMWTAEQALT-------MTGEPAS
MAB_4377c|M.abscessus_ATCC_199 CAKAFKSSDYVAMASQV----MPDAPGLRDVVFIGTSDWTELVAGAERVT
MLBr_02546|M.leprae_Br4923 YDDEYTKVVSKAEPALG---KLRALGVNPDEEEPPGS-----TDETLAEL
Mb3591|M.bovis_AF2122/97 LDAVAARAAAVAPQDVSDILFTSGTTGRSKGVLCAHRQSLSASASWAANG
Rv3561|M.tuberculosis_H37Rv LDAVAARAAAVAPQDVSDILFTSGTTGRSKGVLCAHRQSLSASASWAANG
MUL_4126|M.ulcerans_Agy99 TGAVTARAAAVSPDDVSDILFTSGTTGRSKGVLCAHRQSLSASASWAANG
MAV_1577|M.avium_104 GQIVDAMTQTVTPADPLVIMFTSGSSGTPKGVWHSHGSALGAVQSGLAAR
MMAR_4027|M.marinum_M LRMVEPIAETVTAADTLVIMFTSGSTGAPKGVLHSHGSALGAVRSGLTAR
TH_4352|M.thermoresistible__bu --------------------------------------------------
Mflv_4364|M.gilvum_PYR-GCK AAVSDAMSRGVRAADPMAILFTSGSSGPPKGVIHSHGNAIGAVRSGLDAR
Mvan_1980|M.vanbaalenii_PYR-1 AEVAEAMSRGVRPADPLTVVFTSGSSGAPKGVRHSHGNAIGAVRSGLAAR
MSMEG_2215|M.smegmatis_MC2_155 ---ADALAARVRPSDPMVILFTSGSSGPPKGVIHSHGNAVRAVASGLAAR
MAB_4377c|M.abscessus_ATCC_199 ETALRARMSQLSNTDPINIQYTSGTTGYPKGATLSHRNVLNNGFFVAESI
MLBr_02546|M.leprae_Br4923 IARSSSAPAPKVRKRASIIILTSGTTGTPKGANRNTPASLAPLGGILSKV
Mb3591|M.bovis_AF2122/97 KITSDDRYLCINPFFHNFGYKAGILACLQTGATLIPHVT-FDPLHALRAI
Rv3561|M.tuberculosis_H37Rv KITSDDRYLCINPFFHNFGYKAGILACLQTGATLIPHVT-FDPLHALRAI
MUL_4126|M.ulcerans_Agy99 KITADDRHLCINPFFHNFGYKAGILACLQTGATLFPHLT-FDPLRTLQAI
MAV_1577|M.avium_104 CIDADSRLYLPMPFFWVGGFGSGILSALLAGATLVTEEI-PRPETTLRLL
MMAR_4027|M.marinum_M RITSATRLYLPMPFFWVGGFGGGILSALLAGATLVTEAR-PFPENTLRLL
TH_4352|M.thermoresistible__bu -------LYLPMPFFWVGGFGGGVLSALVAGATLVTEEK-PQPHTTLRLL
Mflv_4364|M.gilvum_PYR-GCK CIDDRTRLYLPMPFFWVGGFGGGLLSALVAGATLVTEAV-PQPDSTLRLL
Mvan_1980|M.vanbaalenii_PYR-1 CVDADTRLYLPMPFFWIGGLGGGLLSALTAGATLITEPV-PQPDSTLRLL
MSMEG_2215|M.smegmatis_MC2_155 CVTADTRLYLPMPFFWVGGFGAGIVTALVAGATLVTEPI-PSPQSTLDLL
MAB_4377c|M.abscessus_ATCC_199 QLQAGDRLCIPVPFYHCFGMVMGNLGCTTHGATIVMPAPGFDPGRTLEAI
MLBr_02546|M.leprae_Br4923 PFRAHEVTLLPAPMFHALGYLHATLAMFLG-STLVLRRR-FKPATVLEDI
*:: * . : :*:. * .* :
Mb3591|M.bovis_AF2122/97 ERHRITVLPGPPTIYQSLLD--HPARKDFDLSSLRFAVTGAATVPVVLVE
Rv3561|M.tuberculosis_H37Rv ERHRITVLPGPPTIYQSLLD--HPARKDFDLSSLRFAVTGAATVPVVLVE
MUL_4126|M.ulcerans_Agy99 ERHRITVLPGPPTIYQTLLD--HPARDDYDLSSLRFAVTGAATVPVVLVE
MAV_1577|M.avium_104 ESERVTLFRGWPDQAETLAR--HAGTVGADLSALRPGSLQALLPPEQRAR
MMAR_4027|M.marinum_M EAERVTMFRGWPDQAEALAR--HPSSVDVDLSALQPGSLPALLPPERRAQ
TH_4352|M.thermoresistible__bu ERERVTLFRGWPDQAEALAR--HRESVGADLSSLRPGSLEALLPPDQRAD
Mflv_4364|M.gilvum_PYR-GCK TAERVTLFRGWPDQAEALAR--HLPASGVELN-LRPGSLEALLPPNLRAR
Mvan_1980|M.vanbaalenii_PYR-1 ADERVTLFRGWPDQAEALAR--HLPASGVELTELRAGSLDALLPPRLRSR
MSMEG_2215|M.smegmatis_MC2_155 HRERVTLFRGWPDQAEALAR--HASST--DLSALRPGSLEALLPHELRAQ
MAB_4377c|M.abscessus_ATCC_199 ERERCVGVYGVPTMFIAMLA--DPGFAHRDLSTLRTGIMAGSVCPVEVMK
MLBr_02546|M.leprae_Br4923 EKHQATAMVVVPVMLSRILDTLENLETKSDLSSLRVVFVSGSQLGAELAT
.: . . * : . :*. *: .
Mb3591|M.bovis_AF2122/97 RMQSELDIDIVLTAYGLTEANGMGTMCRPEDDAVTVAT-TCGRPFADFEL
Rv3561|M.tuberculosis_H37Rv RMQSELDIDIVLTAYGLTEANGMGTMCRPEDDAVTVAT-TCGRPFADFEL
MUL_4126|M.ulcerans_Agy99 RMQSELDIDIVLTAYGLTEANGMGTMCRADDDALTVAT-TCGRPFADFEL
MAV_1577|M.avium_104 -------PGARATLFGMTEAFGPYSGYPADTDMPVSARGSCGKPFDGMEV
MMAR_4027|M.marinum_M -------PGARARLFGMTEAFGPYCGYRADTDMPPTAWGSCGKPFPGMEI
TH_4352|M.thermoresistible__bu -------RGARANLFGMTESFGPYCGYRADTDMPRSAWGSCGKPFPGMEV
Mflv_4364|M.gilvum_PYR-GCK -------PGARANLFGMTESFGPYCGHPADTDMPETAWGSCGTPFDGMQV
Mvan_1980|M.vanbaalenii_PYR-1 -------PGARARLFGMTESFGPYCGYPADTDMPETAWGSCGKPFDGMQV
MSMEG_2215|M.smegmatis_MC2_155 -------PGARASLFGMTESFGPYCGYPADADMPRSAWGSCGRPFDSMDV
MAB_4377c|M.abscessus_ATCC_199 RCIDEMHMAGVAIAYGMTETSPVSCQTLFDDDLDRRTA-TVGRAHPHIEI
MLBr_02546|M.leprae_Br4923 R----ALEELGPVIYNMYGSTEIAFATIAGPKDLQINSATVGPVVKGVKV
:.: : . : * ..:
Mb3591|M.bovis_AF2122/97 RIADD-----------GEVLLRGPNVMVGYLDDTEATAAAIDADGWLHTG
Rv3561|M.tuberculosis_H37Rv RIADD-----------GEVLLRGPNVMVGYLDDTEATAAAIDADGWLHTG
MUL_4126|M.ulcerans_Agy99 RIGPDT----------GEVLLRGPNVMLGYLDDPRATAAAIDADGWLHTG
MAV_1577|M.avium_104 RIVDPDTGAPVGAGTAGIIQIRGPHTLRGMCG--RSREELFTVDGFYPTG
MMAR_4027|M.marinum_M RIVDPETGTRVPAETIGQIQIRGPHTLRGLCR--RSREEVFTADGYYCSG
TH_4352|M.thermoresistible__bu RIVDPDSGAPVPAGTVGEIQLRGPHLMRGICR--RSREEVFTPDGFYPTG
Mflv_4364|M.gilvum_PYR-GCK RITDPGTGEELPAGATGMIQIRGPHLMRGICR--RTREDVFTPDGFYPTG
Mvan_1980|M.vanbaalenii_PYR-1 RIVGADTGEPAPTGAIGMIQIRGRHVMRGICR--RSREEVFTSDGWYATG
MSMEG_2215|M.smegmatis_MC2_155 RIVDTTTGTPLRVDEVGEIQIRGPHILRGICR--RGREEVFTRDGYYPTG
MAB_4377c|M.abscessus_ATCC_199 KIVDPNSGETVQRGQSGELCTRGYSVMLGYWNDEAHTREVLDTDGWMHTG
MLBr_02546|M.leprae_Br4923 KILDDS-GKEVPRGQVGRIFVGNAFPFEGYTG----GGGKQIIDGLLSSG
:* * : . : * ** :*
Mb3591|M.bovis_AF2122/97 DIGAVDQAGNLRINDRLKDMYICGGFNVYPAEVEQVLARMDGVADAAVIG
Rv3561|M.tuberculosis_H37Rv DIGAVDQAGNLRITDRLKDMYICGGFNVYPAEVEQVLARMDGVADAAVIG
MUL_4126|M.ulcerans_Agy99 DIGVLDEAGNLRITDRLKDMYICGGFNVYPAEVEQVLARLDGVADVAVIG
MAV_1577|M.avium_104 DLGHLDDAGFLFYHGRADDMFKVSGATVYPSEVERALRTIDGVDGAVVTN
MMAR_4027|M.marinum_M DLGHLDANGYLFYHGRSDDMFKVSGATVYPVEVETALRTIDGVSNAFVTN
TH_4352|M.thermoresistible__bu DLGRLDADGFLFYHGRRDDMFKVSGATVYPGEVADALRSIDGVRAAYVTD
Mflv_4364|M.gilvum_PYR-GCK DLGHLDSDGFLFYHGRCDDMFKVRGATVYPAEVEQALRTVDGVRAASVTN
Mvan_1980|M.vanbaalenii_PYR-1 DLGHLDADGFLYYHGRCDDMFKVRGATVYPSEVEQALRSLDGVSAAFVTN
MSMEG_2215|M.smegmatis_MC2_155 DLGRLDSDGFLFYHGRSDDMFKVSGATVYPGEAQHALRGIAGVRAAYVVN
MAB_4377c|M.abscessus_ATCC_199 DLAVMRDDGYCTIIGRLKDMVIRGGENIYPREIEEFLLTHPDIEDVHVVG
MLBr_02546|M.leprae_Br4923 DVGYFDEHDLLYISGRDDEMIVSGGENVFPAEVEDLISGHPEVVEATAIG
*:. . . .* .:* * .::* * : : . . .
Mb3591|M.bovis_AF2122/97 VPDQRLGEVGRAFVVARPG-TGLDEASVIAYTREHLANFKTPRSVRFVD-
Rv3561|M.tuberculosis_H37Rv VPDQRLGEVGRAFVVARPG-TGLDEASVIAYTREHLANFKTPRSVRFVD-
MUL_4126|M.ulcerans_Agy99 IPDHRLGEVGRAFIVARPG-FNLDEKSVIDYTREHLANFKAPRSVRFVD-
MAV_1577|M.avium_104 VP-GATGDRVGAAVVCR----ELTAAQLRAAARNLLSSFKVPSVWLVLRS
MMAR_4027|M.marinum_M VA-GAQGQRVAAAVVCST--PALTSEDLRTSAGQVLSAFKVPTVWLLLAD
TH_4352|M.thermoresistible__bu VADGRGGDRVGAAVVGT----VTDVGQLRREARKLLSSFKVPTVWLLLDS
Mflv_4364|M.gilvum_PYR-GCK LP-GPEGNRVGAAVVCD---ASLTVDVLRRAAREVLSSFKVPSVWALLPD
Mvan_1980|M.vanbaalenii_PYR-1 VA-GPEGDRVGAAVVCD---APATAERLRSAAREVLSSFKVPTVWLLLDS
MSMEG_2215|M.smegmatis_MC2_155 VP-DHQRNRVGAVVVTD----GMSARDLCAAAREVLSPFKVPSVWALLDD
MAB_4377c|M.abscessus_ATCC_199 VPDEKYGEELCAWVRMRPDRVVIDAVAIRAFASGRLAHYKIPRYVHVVES
MLBr_02546|M.leprae_Br4923 VDDKEWGARLRAFVVKKQD-ATIDEDAIKAYVRDHLARYKVPREVIFLE-
: * : : . *: :* * .:
Mb3591|M.bovis_AF2122/97 --VLPRNAAGKVSKPQLRELG-------------
Rv3561|M.tuberculosis_H37Rv --VLPRNAAGKVSKPQLRELG-------------
MUL_4126|M.ulcerans_Agy99 --TLPRNAGGKVVKPQLRELA-------------
MAV_1577|M.avium_104 DDDLPRGVTGKVDVRRLRELLADADRRQETRVQG
MMAR_4027|M.marinum_M DANIPRGPTGKVDLQRLRDMLANTA---------
TH_4352|M.thermoresistible__bu DDAVPRRATGKVDVNRLRDLLATHPDRVVT----
Mflv_4364|M.gilvum_PYR-GCK EAAIPRGATGKVDTARLRELLQD-----------
Mvan_1980|M.vanbaalenii_PYR-1 QDGVPRGATGKVDVTRLRALLEQS----------
MSMEG_2215|M.smegmatis_MC2_155 EAAVPIGATGKVDNNGLRAMLAERGER-------
MAB_4377c|M.abscessus_ATCC_199 FPMTVTGKVRKIEMRQQTIQIFGLPEPGRTDEQ-
MLBr_02546|M.leprae_Br4923 --ELPRNPTGKVLKRELRDFEVH-----------
*: