For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTMNAKRRRMQQRLAKLPGVRAVRRPISPDTADEFDLYYVRAGRKSRHPVVIIPGGPGVASVQMYKGLRR RAAAAGLDVIMIEHRGVGMSRHDDAGADLPPEAMTVNQVVDDVAAVLDDARVDSAVVYGASYGTYIASGV GVRHPARVHGMVLDSPLLSRHDIVIVRRAIRRLLLRGDSPETASLAPKMRKLVDAGLMTATATQVVATIY GYGGARLLERQLDLLLDGRKLLWWTLSRFTTLRNLPFRYEEDLVSRIALRELDYAAEPDGLPLDPAVAER EVRTQTATFEDEPYDLPAEMPSFDWPTAVVSGGRDLITPPAVAERVASLLPHAVLLSLPDMAHSALDFHE AVALASADAVLRGDLAGLAARACTLNALPPRPEVRLLWALIAAAAMIEGALPIPAPLRRRLSCG
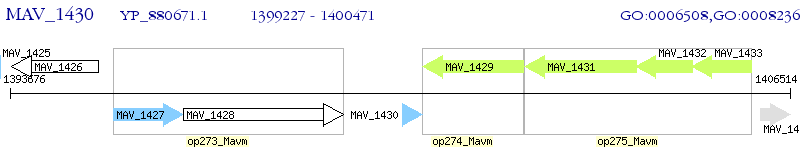
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1430 | - | - | 100% (414) | hydrolase, alpha/beta fold family protein |
| M. avium 104 | MAV_4515 | - | 4e-05 | 27.71% (166) | hydrolase, alpha/beta fold family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2257 | - | 1e-127 | 57.46% (409) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01899 | lipG | 2e-07 | 26.95% (141) | putative hydrolase |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4140 | - | 1e-162 | 70.29% (414) | hydrolase |
| M. smegmatis MC2 155 | MSMEG_4998 | - | 1e-133 | 62.00% (400) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_1861 | lipG | 1e-08 | 31.30% (131) | PROBABLE LIPASE/ESTERASE LIPG |
| M. ulcerans Agy99 | MUL_0733 | lipG1 | 9e-06 | 27.33% (150) | lipase/esterase LipG1 |
| M. vanbaalenii PYR-1 | Mvan_4434 | - | 1e-121 | 55.88% (417) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2257|M.gilvum_PYR-GCK MSVMTMNARRRRVHDRLAALPGVRPVRRPVHPG---ADGHFDVFYVRTGR
Mvan_4434|M.vanbaalenii_PYR-1 MRVMTMNGRRRRAHDKLAALPGVRPVRRPVRPGVGDADEKFDVYYVRTGR
MAV_1430|M.avium_104 ---MTMNAKRRRMQQRLAKLPGVRAVRRPISPD---TADEFDLYYVRAGR
MMAR_4140|M.marinum_M ---MTMSDKRRRAHEKLAGMPGVRPVRRPVSPG---NDSEFDLYYVRAGR
MSMEG_4998|M.smegmatis_MC2_155 ------MANHGRKHATLAALPGVRAVRRPVAPG---ADEQFDVFYVRAGR
MLBr_01899|M.leprae_Br4923 ----------MEVRSGYATS------------------GDLKLYYEDMGN
MUL_0733|M.ulcerans_Agy99 ----------MEIRTGYAQSGVGSGATSERQPA-----GDLKLYYEDMGD
TH_1861|M.thermoresistible__bu ----------MQIRTGFANVD---------GSP-----EPVDLYYEDLGD
. : * ..::* *
Mflv_2257|M.gilvum_PYR-GCK KSAHPLVVIPGGPGAASIALYRSFRRHAAVAGLDVIMVEHRGVGLSRHDD
Mvan_4434|M.vanbaalenii_PYR-1 KSAHPLVIIPGGPGAASIALYRAFRRRAAVAGLDVIMVEHRGVGLSRHDD
MAV_1430|M.avium_104 KSRHPVVIIPGGPGVASVQMYKGLRRRAAAAGLDVIMIEHRGVGMSRHDD
MMAR_4140|M.marinum_M KSAHPLVIIPGGPGVASMLVYKGLRRRAAAAGLDVIMIEHRGVGMSRHDD
MSMEG_4998|M.smegmatis_MC2_155 KSRHPVVLIPGGPGVASIQQYRGLRRRLATSGLDVIMVEHRGIGMSRRTD
MLBr_01899|M.leprae_Br4923 VDDPPVLLIMG-LGAQLVLWRTAFCEKLVAQGLRVVRYDNRDVGLSSRTD
MUL_0733|M.ulcerans_Agy99 INAPPVLLIMG-LGAQLLLWRTGFCQKPVDQGLRVIRYDNRDVGLSTKME
TH_1861|M.thermoresistible__bu PADPPVLLIMG-LGAQLILWRDGFCRKLVERGLRVIRFDNRDVGLSSKLD
*:::* * *. : .: .: . ** *: ::*.:*:* : :
Mflv_2257|M.gilvum_PYR-GCK D------------------GADLPPEALTVDAAVGDIAAVLDDAGVERAV
Mvan_4434|M.vanbaalenii_PYR-1 S------------------GADLPPEALTVDAAVDDIAAVLDDAQVERAS
MAV_1430|M.avium_104 A------------------GADLPPEAMTVNQVVDDVAAVLDDARVDSAV
MMAR_4140|M.marinum_M S------------------GADLPPEAFTIDQVVDDIAAVLDDAHVDSAV
MSMEG_4998|M.smegmatis_MC2_155 A------------------GADLPAEAITVEQVLDDIAAVLDDAGVGTAV
MLBr_01899|M.leprae_Br4923 STEQPCPSQPLTARLIRFWLGQRNACAYTLEDMTDDAVALLDHLSIERAH
MUL_0733|M.ulcerans_Agy99 ---QPSARQPLMPQLIRSWLGRRSQTPYTLEDMADDAAALLDHLDIERAH
TH_1861|M.thermoresistible__bu ---GRRERTPLPWRMLRSFAGLPSPAVYTLEDMADDAAGLLDHLQIDAAH
. *:: .* ..:**. : *
Mflv_2257|M.gilvum_PYR-GCK VYGASYGSYLAAGVGVRHPGRIHAMVLDSPLLSAHDIDAVRAQTRRVLWE
Mvan_4434|M.vanbaalenii_PYR-1 VYGTSYGSYLAAGVGVRHPGRVQSMVLDSPLLSADDIDAVRAQTRRVLWD
MAV_1430|M.avium_104 VYGASYGTYIASGVGVRHPARVHGMVLDSPLLSRHDIVIVRRAIRRLLLR
MMAR_4140|M.marinum_M IYGTSYGTYIAAGVGVRHPARVRSMILDSPLLSRDDIALARETIRRRLLD
MSMEG_4998|M.smegmatis_MC2_155 VYGTSYGSYLASGVGVRHPGRVAAMVLDSPVLSAHDLDEMRTAIRNLLWE
MLBr_01899|M.leprae_Br4923 IVGASMGGMIAQIFAARFPTRTRSLAV---FFSSNNRPFLPPP-------
MUL_0733|M.ulcerans_Agy99 IVGASMGGMIAQIFAARFAERTNSLAV---IFSSNNRPFLPPP-------
TH_1861|M.thermoresistible__bu IVGASMGGMVAQVFAGRHAHRTRSLGV---IFSSNNRPFLPPP-------
: *:* * :* .. *.. * .: : .:* .:
Mflv_2257|M.gilvum_PYR-GCK D-----TDLAPRVRTLADEGALTPQDVLLMIGLYELAGPQVLRRQLDLRV
Mvan_4434|M.vanbaalenii_PYR-1 G-----ADLAPRMRRLAADGVLRSEDVLLAIGLYELAGPEGLGRQLDLLL
MAV_1430|M.avium_104 GDSPETASLAPKMRKLVDAGLMTATATQVVATIYGYGGARLLERQLDLLL
MMAR_4140|M.marinum_M GDSPETAALAAKVRQLVDAGVMTARASQVAAAVYGYGGARLLERQLDLLL
MSMEG_4998|M.smegmatis_MC2_155 GTGPDTAALAPKVRKLVETGRLTG--GQLAGLLYGYGGARLLDRQLDLLL
MLBr_01899|M.leprae_Br4923 ---------APRALLALLTGPPTG--------------------------
MUL_0733|M.ulcerans_Agy99 ---------APRALLAVLNGPPPG--------------------------
TH_1861|M.thermoresistible__bu ---------GPRQLLSIITGPAPD--------------------------
..: *
Mflv_2257|M.gilvum_PYR-GCK ADRNRLWKIAERVTRLVTERKTPYHHEPDLVERIAYQELNYGAVPDGGPV
Mvan_4434|M.vanbaalenii_PYR-1 ARRDWLWRVAESVTKLVTERKTAYRHEPDLVERIAYRELNYGAVPDGGPL
MAV_1430|M.avium_104 DGRKLLWWTLSRFTTL---RNLPFRYEEDLVSRIALRELDYAAEPDGLPL
MMAR_4140|M.marinum_M CGRMILWHVLFRFSAMY-ARKVPYRYEMDLVGRIAFRELDYAAEPDGLPL
MSMEG_4998|M.smegmatis_MC2_155 DGHTLLWRTLGRLGRVS-TRKAPYRNEIDLVGRIAFRELNYAGVPDGLPL
MLBr_01899|M.leprae_Br4923 SRRDVVVDNVVRVTKIT--GSPLYRMPEEQVRTNAAEIYDRSFYPLGVSR
MUL_0733|M.ulcerans_Agy99 SPREVIIDNVVRVTRIT--GSPGYQPTEEWIRADAAENYDRSYYPWGIPR
TH_1861|M.thermoresistible__bu APRDEVIENAVKVGRLN--GSPAYPVPDEQLRADAATAFDRSYHPVGVAR
: : . : . : : : * : . * * .
Mflv_2257|M.gilvum_PYR-GCK DPAVALREMASGT---VEFVGEPYDLPAAMPGFGWPTAVLSGGRDLTTPP
Mvan_4434|M.vanbaalenii_PYR-1 DPSVALRQMATGD---VDFVAEPYDLTSEMPGFAWPTVVVSGGRDLTTPP
MAV_1430|M.avium_104 DPAVAEREVRTQT---ATFEDEPYDLPAEMPSFDWPTAVVSGGRDLITPP
MMAR_4140|M.marinum_M DPAQAIRETIAAT---DQFESEPYDLVAEMPRFGWPTVVVSGGHDLTTPA
MSMEG_4998|M.smegmatis_MC2_155 DPALEMAEMARATKFHEPFEAEPYDLTAEMPKFTWPTVVVSGGRDLITPP
MLBr_01899|M.leprae_Br4923 QFSAILGSGSLLH----------YNQRIIAP-----TVVIHGRADKLVRP
MUL_0733|M.ulcerans_Agy99 QFSAVLGSGSLLG----------YNRRTVAP-----TVVIHGRADKLMRP
TH_1861|M.thermoresistible__bu QFDAILGSGSLRR----------YNRRTTSP-----TVVIHGRADKLMRP
: . *: * *.*: * * .
Mflv_2257|M.gilvum_PYR-GCK EVARRVADLIPDSVLVSLPTAGHSVLDIRERAAVEICKAVCSGELHELPS
Mvan_4434|M.vanbaalenii_PYR-1 EVARRIADLIPDAALLELPTAGHSILDTREHAALRICEAVQAGQSHDLPS
MAV_1430|M.avium_104 AVAERVASLLPHAVLLSLPDMAHSALDFHEAVALASADAVLRGDLAGLAA
MMAR_4140|M.marinum_M MVAEHIASLLPNAVLIELPTMAHSALDFREPAALAIAGAVCRGEFGALTG
MSMEG_4998|M.smegmatis_MC2_155 ALAQRVASLIPGATPVCMATAAHSIVDTRERAAIAIMQAVCNGAAEQLPH
MLBr_01899|M.leprae_Br4923 SSGRAVARAITGARLVLFDGMGHDLPQQLWDQAVGVLMSNFAKAS-----
MUL_0733|M.ulcerans_Agy99 FGGRAVAKAINGARMVEFDGMGHDLPRQLWDQVIGVLTSNFEQAE-----
TH_1861|M.thermoresistible__bu TGGRAVARAIPGARLTLFDGMGHDLPEPLWDEIVAELDKNITAGR-----
.. :* : : : .*. :
Mflv_2257|M.gilvum_PYR-GCK RAAELDALPANLTVRLLVGAITVAARAESAVPGSAP----TAPTS
Mvan_4434|M.vanbaalenii_PYR-1 RSTELDALPANLTVRLLVRAITVAARAESAVPGAIPRVVRQLATS
MAV_1430|M.avium_104 RACTLNALPPRPEVRLLWALIAAAAMIEGALPIPAPLRRRLSCG-
MMAR_4140|M.marinum_M RGEELDAMPARLPIRALWKAIELAAMAEHALPAAPLLHRRVSSA-
MSMEG_4998|M.smegmatis_MC2_155 TADELDALPPRMPIRLLVAAISAAVAAEGMADSVLPATS------
MLBr_01899|M.leprae_Br4923 ---------------------------------------------
MUL_0733|M.ulcerans_Agy99 ---------------------------------------------
TH_1861|M.thermoresistible__bu ---------------------------------------------