For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EPISPTDALFLIGESREHPMHVGSLQLFEPPPDAGPHFVRESYQAMLECTDVQPTFRKHPAFFGGVTNLA WSIDKEVELDYHLRRSALPEPGRVRDLLELASRLHGSLLDRHRPLWEAHLVEGLQDGRYAVYTKYHHSLM DGVSALRLVQRAFTPDPDDDEVRVPWSLAPRQRGPRRRPSLLERAGRTAGSALALAPSTLKLARAALLEQ QLTLPFQAPRSMFNVRIGGARRVAAQSWPLERINAVKAATGATVNDVILAMSSGALRAYLLDQNALPDAP LTAMVPVNLRKPDDDRGGNLVGTFLCNLATDLDDPAKRLEIISASIRDTKEVFWQLPPVQQLALSAFNIG GLFFGLIPGYLSTASPPFNIVISNVSTGRSERLYWRGARLDGNYPLSIPLDGQAVNITVTNNADNLDFGL VGCRRSVPHLQRMLGHLEASLKELELAAGV*
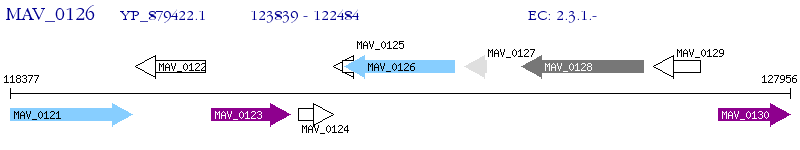
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0126 | - | - | 100% (451) | bifunctional wax ester synthase/acyl-CoA diacylglycerol acyltransferase |
| M. avium 104 | MAV_0355 | - | e-172 | 67.84% (454) | bifunctional wax ester synthase/acyl-coadiacylglycerol |
| M. avium 104 | MAV_3352 | - | 1e-53 | 34.53% (472) | acyltransferase, ws/dgat/mgat subfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3761c | - | 1e-164 | 63.97% (458) | hypothetical protein Mb3761c |
| M. gilvum PYR-GCK | Mflv_1058 | - | 3e-77 | 36.97% (468) | hypothetical protein Mflv_1058 |
| M. tuberculosis H37Rv | Rv3734c | - | 1e-164 | 63.97% (458) | hypothetical protein Rv3734c |
| M. leprae Br4923 | MLBr_01244 | - | 1e-20 | 28.47% (281) | hypothetical protein MLBr_01244 |
| M. abscessus ATCC 19977 | MAB_4544c | - | 1e-121 | 48.14% (457) | hypothetical protein MAB_4544c |
| M. marinum M | MMAR_1809 | - | 0.0 | 74.17% (453) | hypothetical protein MMAR_1809 |
| M. smegmatis MC2 155 | MSMEG_6322 | - | 1e-171 | 65.42% (454) | bifunctional wax ester synthase/acyl-CoA diacylglycerol |
| M. thermoresistible (build 8) | TH_2030 | - | 1e-162 | 63.80% (453) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2057 | - | 0.0 | 74.17% (453) | hypothetical protein MUL_2057 |
| M. vanbaalenii PYR-1 | Mvan_5773 | - | 1e-75 | 37.15% (463) | hypothetical protein Mvan_5773 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1058|M.gilvum_PYR-GCK MSANSVRLGLQDLM----FIYGETSSSKMHVGGLLPFTPP-ADAPRDYLR
Mvan_5773|M.vanbaalenii_PYR-1 -------------M----FIYGETSSSKMHVAGLLPFTPP-ADAPRDYLR
Mb3761c|M.bovis_AF2122/97 ----MDLMMPNDSM----FLFIESREHPMHVGGLSLFEPP-QGAGPEFVR
Rv3734c|M.tuberculosis_H37Rv ----MDLMMPNDSM----FLFIESREHPMHVGGLSLFEPP-QGAGPEFVR
MMAR_1809|M.marinum_M ----MKLIAPTDSM----FLLGESREHPMHVGGLQLFQPP-EDADENFVT
MUL_2057|M.ulcerans_Agy99 ----MKLIAPTDSM----FLLGESREHPMHVGGLQLFQPP-EDADENFVT
MAV_0126|M.avium_104 ----MEPISPTDAL----FLIGESREHPMHVGSLQLFEPP-PDAGPHFVR
MSMEG_6322|M.smegmatis_MC2_155 -MNRMQLMSPTDSM----FLIAESREHPMHVGGLALYDPP-DDAGPEFVR
TH_2030|M.thermoresistible__bu ----MRLISPADSM----FLLAESREHPMHVAGLQLFEPPPGTAGPEFLQ
MAB_4544c|M.abscessus_ATCC_199 ----MPFMPVSDSM----FLIGETREHPFHVGGLQLFKPP-VDAGPDYAE
MLBr_01244|M.leprae_Br4923 MADSVEGIGPFDELGALDYLLHRGEANPRTRAGIMAVELL--DTTPDWN-
: :: . ..: : .:
Mflv_1058|M.gilvum_PYR-GCK GMIDEARRQEVVAPWNRRLAYPRLQFSPLHSWVTDEDFDFDYHVRRSALA
Mvan_5773|M.vanbaalenii_PYR-1 RMVDDARRQEVMPPWNRKLAYPRLQFSPLHTWVTDTEFDFDYHVRRSALA
Mb3761c|M.bovis_AF2122/97 EFTERLVANDEFQPMFRKHPATIGGGIARVAWAYDDDIDIDYHVRRSALP
Rv3734c|M.tuberculosis_H37Rv EFTERLVANDEFQPMFRKHPATIGGGIARVAWAYDDDIDIDYHVRRSALP
MMAR_1809|M.marinum_M EAYEAMLGCTDVQPMFRKHPA-FLGAMTNFAWTFDDDVELDYHFRRSALP
MUL_2057|M.ulcerans_Agy99 EAYEAMLGCTDVQPMFRKHPA-FLGAMTNFAWTFDDDVELDYHFRRSALP
MAV_0126|M.avium_104 ESYQAMLECTDVQPTFRKHPA-FFGGVTNLAWSIDKEVELDYHLRRSALP
MSMEG_6322|M.smegmatis_MC2_155 ELYEEMVRHTDFQPVFRKHPATLLGGIANVGWTLDDEVDLDYHLRRSALP
TH_2030|M.thermoresistible__bu GLHQRLAAGRELHPTFRKHPATLFGGIAHLGWAVDDDVDFDYHLRRSALP
MAB_4544c|M.abscessus_ATCC_199 VFYEQLMSTTEVSSDFRKRPGQPLAVMGNLTWIVDDEVDWEYHVRRSALP
MLBr_01244|M.leprae_Br4923 RFRSRIEDVSQRVLRLRQKVVVPTLPTAAPRWVVDPDFELDFHVRRVRVP
. *: * * :.: ::*.** :.
Mflv_1058|M.gilvum_PYR-GCK SPGDERELGVLVSRLHSNHLDLTRPPWELHVIEGLEGGRFALYMKIHHAL
Mvan_5773|M.vanbaalenii_PYR-1 SPGDERELGILVSRLHSNHLDLTRPPWELHVIEGLEGGRFALYMKIHHAL
Mb3761c|M.bovis_AF2122/97 SPGRVRDLLELTSRLHTSLLDRHRPLWELHVVEGLNDGRFAMYTKMHHAL
Rv3734c|M.tuberculosis_H37Rv SPGRVRDLLELTSRLHTSLLDRHRPLWELHVVEGLNDGRFAMYTKMHHAL
MMAR_1809|M.marinum_M RPGRVRELLELTSRLHGSLLDRHRPLWEAHLVEGLNDGRYAVYIKFHHSL
MUL_2057|M.ulcerans_Agy99 RPGRVRELLELTSRLHGSLLDRHRPLWEAHLVEGLNDGRYAVYIKFHHSL
MAV_0126|M.avium_104 EPGRVRDLLELASRLHGSLLDRHRPLWEAHLVEGLQDGRYAVYTKYHHSL
MSMEG_6322|M.smegmatis_MC2_155 SPGRPRELLELTSRVHGTLLDRHRPLWEAYLIEGMADGRFAVYTKVHHSL
TH_2030|M.thermoresistible__bu GPGRIRELLDLTSRWHGTLLDRHRPLWEVHLVEGLADGRVAIYFKVHHAL
MAB_4544c|M.abscessus_ATCC_199 RPARVRELLTVTSRWHSSPLDRHRPLWEMHVVEGLSDGRLAVYTKMHHAV
MLBr_01244|M.leprae_Br4923 DPGTLREVFDLAEVIQQSPMDVSRPLWTATLVEGLAAGRAAMLLQISHAI
*. *:: :.. : . :* ** * ::**: ** *: : *::
Mflv_1058|M.gilvum_PYR-GCK VDGYSAMRMLGRSLSTDPASRDTRMFFNVPSPTRSRRDPGAAESSNPLTA
Mvan_5773|M.vanbaalenii_PYR-1 VDGYTAMRMLGRSLSTDPETRDARMFFNVPIPKRSRPTGAGAQSSNPVTA
Mb3761c|M.bovis_AF2122/97 IDGVSAMKLAQRTLSADPDDAEVRAIWNL--PPRPRTRPPSDGSS-----
Rv3734c|M.tuberculosis_H37Rv IDGVSAMKLAQRTLSADPDDAEVRAIWNL--PPRPRTRPPSDGSS-----
MMAR_1809|M.marinum_M QDGVSAQKLMQRAFSTDPDDDEIRVPWEL--KPR-RRSNPGHRSS-----
MUL_2057|M.ulcerans_Agy99 QDGVSAQKLMQRAFSTDPDDDEIRVPWEL--KPR-RRSNPGHRSS-----
MAV_0126|M.avium_104 MDGVSALRLVQRAFTPDPDDDEVRVPWSL--APR-QRG-PRRRPS-----
MSMEG_6322|M.smegmatis_MC2_155 IDGVSAMKLVERTLSEDPSDTTVRVPWNL--PRR-ESSRRAGSSS-----
TH_2030|M.thermoresistible__bu IDGVAVMKLMQRTLSTDPGDD-ARVPWNL--PPP-RRNP-AGPVS-----
MAB_4544c|M.abscessus_ATCC_199 IDGVGALRMMMRSLSDDPDARDCQAVWAP--AKRRAKSSIVSTTNTSAID
MLBr_01244|M.leprae_Br4923 TDGVGSVEMFAEMYDLERDPPSRPRSPQPIPQDLSRNDLMLQGINHLPVA
** .: . : .
Mflv_1058|M.gilvum_PYR-GCK TLRALGGVSSAVTGGVSSAVDLTNALVNTQIRRDGENAHIAGSVSAPHSI
Mvan_5773|M.vanbaalenii_PYR-1 TLRALGAVGSTVSDGVGSAVDLASALVNTQIRRDGDFGRISGSASAPHSI
Mb3761c|M.bovis_AF2122/97 LLDALFKMAGSVVGLAPSTLKLARAALLEQQ--------LTLPFAAPHSM
Rv3734c|M.tuberculosis_H37Rv LLDALFKMAGSVVGLAPSTLKLARAALLEQQ--------LTLPFAAPHSM
MMAR_1809|M.marinum_M PLRLLTETVGATASLAPSALSLARAALLEQQ--------LTLPFRAPKTM
MUL_2057|M.ulcerans_Agy99 PLRLLTETVGATASLAPSALSLARAALLEQQ--------LTLPFRAPKTM
MAV_0126|M.avium_104 LLERAGRTAGSALALAPSTLKLARAALLEQQ--------LTLPFQAPRSM
MSMEG_6322|M.smegmatis_MC2_155 LARTATGAATSLAALAPSTIRLARAALLEQQ--------LTLPFGAPRTM
TH_2030|M.thermoresistible__bu RLRSATGAVGSVAALAPSTLSLARAALLEQR--------LTLPFGAPRTM
MAB_4544c|M.abscessus_ATCC_199 LVKGAAGAVRQVVSIPPGLLKYGRHALSDPQ--------LVKPLSAPHTM
MLBr_01244|M.leprae_Br4923 LAGGVVGGLSGVASVAGRAILRPASTVSGVVGYVRSGIRVLSQAAEPSPL
: : : * .:
Mflv_1058|M.gilvum_PYR-GCK LNARISRNRRFATQQYEFDRLKKLSSQHGATLNDVALAIIGGGLRKFLSD
Mvan_5773|M.vanbaalenii_PYR-1 LNARISRNRRFATQQYEFDRLKKLSSQHGATINDVALAIIGGGLRKFLAD
Mb3761c|M.bovis_AF2122/97 FNVKVGGARRCAAQSWSLDRIKSVKQAAGVTVNDAVLAMCAGALRYYLIE
Rv3734c|M.tuberculosis_H37Rv FNVKVGGARRCAAQSWSLDRIKSVKQAAGVTVNDAVLAMCAGALRYYLIE
MMAR_1809|M.marinum_M FNVRIGGARRVAAQSWSLDRIKAVKNAADVTVNDVVLAMSAGALRAYLID
MUL_2057|M.ulcerans_Agy99 FNVRIGGARRVAAQSWSLDRIKAVKNAADVTVNDVVLAMSAGALRAYLID
MAV_0126|M.avium_104 FNVRIGGARRVAAQSWPLERINAVKAATGATVNDVILAMSSGALRAYLLD
MSMEG_6322|M.smegmatis_MC2_155 FNVKIGGARRVAAQSWPLERLRRIKAVTGATINDIVLAMCAGALRAYLAE
TH_2030|M.thermoresistible__bu FNVRIGGARRVAAQSWPLQRIRRIRQAAGXTLNDVALAMCAGALRQYLLE
MAB_4544c|M.abscessus_ATCC_199 LNVSIGGARRFAAQTWSMARIKEAGKALGGTVNDVVLAMCGGALRTYLEE
MLBr_01244|M.leprae_Br4923 LRQRS-LATRTEAIEIQLSDLHKAAKAGDGSINDAYLAGLVGALRRYH-E
:. * : : :. . ::** ** *.** : :
Mflv_1058|M.gilvum_PYR-GCK FDKLPDRSLIAFLPVNVRPKGDEGGG-NAVGAILAP-MGTDIEDPVERLD
Mvan_5773|M.vanbaalenii_PYR-1 LGELPDRSLIAFLPVNVRPKGDEGGG-NAVGAILAP-MGTDIEDPVERLA
Mb3761c|M.bovis_AF2122/97 RNALPDRPLIAMVPVSLRSKEDADAGGNLVGSVLCN-LATHVDDPAQRIQ
Rv3734c|M.tuberculosis_H37Rv RNALPDRPLIAMVPVSLRSKEDADAGGNLVGSVLCN-LATHVDDPAQRIQ
MMAR_1809|M.marinum_M QHALPDAPLIAMVPVSLRKEGDADRGGNMVGTLLCN-LATNLDDPMKRLQ
MUL_2057|M.ulcerans_Agy99 QHALPDAPLIAMVPVSLRKEGDADRGGNMVGTLLCN-LATNLDDPMKRLQ
MAV_0126|M.avium_104 QNALPDAPLTAMVPVNLRKP-DDDRGGNLVGTFLCN-LATDLDDPAKRLE
MSMEG_6322|M.smegmatis_MC2_155 QDALPDRPLIAMVPVSMRSEHEADAGGNMVGSILCS-LGTDVEDPADRLA
TH_2030|M.thermoresistible__bu HDALPATPLVAMVPVSLRSADESAGGGNRTGLVLCN-LATDSDDPAERLD
MAB_4544c|M.abscessus_ATCC_199 QNALPDKPLVAMCPVSIRAEGDQNAG-NSITALLAN-LATDKPDPLERFS
MLBr_01244|M.leprae_Br4923 ALGVSISTLPMAVPVNVRTEADVVGSNRFVGVTLAAPLGTN--DPAARMQ
:. .* **.:* : . . *. :.*. ** *:
Mflv_1058|M.gilvum_PYR-GCK TITTATRAAKGQLQSMSPAAIIAYSAALLAPAGSQIAGALTGVQPPWPYT
Mvan_5773|M.vanbaalenii_PYR-1 VITAATRASKAQLQSMSPAAIIAYSAALLAPAGGQIAGALTGVNPPWPHT
Mb3761c|M.bovis_AF2122/97 TISASMDGNKKVLSELPQLQVLALSALNMAPL--TLA-GVPGFLSAVPPP
Rv3734c|M.tuberculosis_H37Rv TISASMDGNKKVLSELPQLQVLALSALNMAPL--TLA-GVPGFLSAVPPP
MMAR_1809|M.marinum_M VISQSMSDNKKVFAQLPRIQQLALSGLLTGGL--VFG-LVPGFVATAPPP
MUL_2057|M.ulcerans_Agy99 VISQSMSDNKKVFAQLPRIQQLALSGLLTGGL--VFG-LVPGFVATAPPP
MAV_0126|M.avium_104 IISASIRDTKEVFWQLPPVQQLALSAFNIGGL--FFG-LIPGYLSTASPP
MSMEG_6322|M.smegmatis_MC2_155 VIRRSITDNKRVFSELPRLQALALSALLIAPL--GLTTAFPGFVDATAPP
TH_2030|M.thermoresistible__bu RISSSMRSNKRVFSQLPRVQAMALSAAMIAPL--GLG-AVPGVVSSAPPP
MAB_4544c|M.abscessus_ATCC_199 AIRDSVQAAKSVLSEMTPLQRMLVGALNGAPV--LTG-AVPGLVDRTAPA
MLBr_01244|M.leprae_Br4923 KIRSQMTQRR----DEPAMNIIGS----LAPLMTVLPASVLDFIVDSVAS
* : . . : . . . .
Mflv_1058|M.gilvum_PYR-GCK FNLCVSNVP-GPREPLYFNGSRLEATYPVS----------IPIHGMALNI
Mvan_5773|M.vanbaalenii_PYR-1 FNLCVSNVP-GPREPLYFNGSRLEATFPVS----------IPIHGMALNI
Mb3761c|M.bovis_AF2122/97 FNIVISNVP-GPVDPLYYGTARLDGSYPLS----------NIPDGQALNI
Rv3734c|M.tuberculosis_H37Rv FNIVISNVP-GPVDPLYYGTARLDGSYPLS----------NIPDGQALNI
MMAR_1809|M.marinum_M FNVVISNVP-GAPNPLYWRGARLVGNYPLS----------IALDGQAMNI
MUL_2057|M.ulcerans_Agy99 FNVVISNVP-GAPNPLYWRGARLVGNYPLS----------IALDGQAMNI
MAV_0126|M.avium_104 FNIVISNVSTGRSERLYWRGARLDGNYPLS----------IPLDGQAVNI
MSMEG_6322|M.smegmatis_MC2_155 FNLVISNVP-GPKKPLYWRGARLAGNYPLS----------IALDGQALNM
TH_2030|M.thermoresistible__bu FNLVISNVP-GAPEPRYWMGARLAGNYPLS----------IVLDGQALNI
MAB_4544c|M.abscessus_ATCC_199 FNVIISNVP-GPRTDMFWNGFEMDGCYPAS----------IPIDGVALNI
MLBr_01244|M.leprae_Br4923 SDVNASNIP-AYPGDTYFAGAKILRQYGIGPRPGVAMMAVLMSRGGFCTV
:: **:. . :: .: : . * .:
Mflv_1058|M.gilvum_PYR-GCK TLQSYADTMNLGFVGCRDRLPHLQRLAVYTGDALAELEAASQI-------
Mvan_5773|M.vanbaalenii_PYR-1 TLQSYADTMNLGFVGCRDRLPRLQRLAVYTGDALGELEAASS--------
Mb3761c|M.bovis_AF2122/97 TLVNNAGNLDFGLVGCRRSVPHLQRLLAHLESSLKDLEQAVGI-------
Rv3734c|M.tuberculosis_H37Rv TLVNNAGNLDFGLVGCRRSVPHLQRLLAHLESSLKDLEQAVGI-------
MMAR_1809|M.marinum_M TVTNNADNLDFGLVGCRRSVPRLQRMLGHLETSLKELERAVGV-------
MUL_2057|M.ulcerans_Agy99 TVTNNADNLDFGLVGCRRSVPRLQRMLGHLETSLKELERAVGV-------
MAV_0126|M.avium_104 TVTNNADNLDFGLVGCRRSVPHLQRMLGHLEASLKELELAAGV-------
MSMEG_6322|M.smegmatis_MC2_155 TVVSNAHNLDFGLVGCRRSVPHLQRLLGHLETSLKDLETATGA-------
TH_2030|M.thermoresistible__bu TLVTNADNLDFGLVGARGSVPHLQRLLGHLEDSLQQLERAVGG-------
MAB_4544c|M.abscessus_ATCC_199 TCTSSGSSMHFGLTGCRTSVPHLQRILAHLEDALTQLEESVA--------
MLBr_01244|M.leprae_Br4923 TVRYDRASVKSEALFARCLLEGFDEVLALAGDPTPHAVPASFAARSSGSP
* .:. .* : ::.: . . :
Mflv_1058|M.gilvum_PYR-GCK -------
Mvan_5773|M.vanbaalenii_PYR-1 -------
Mb3761c|M.bovis_AF2122/97 -------
Rv3734c|M.tuberculosis_H37Rv -------
MMAR_1809|M.marinum_M -------
MUL_2057|M.ulcerans_Agy99 -------
MAV_0126|M.avium_104 -------
MSMEG_6322|M.smegmatis_MC2_155 -------
TH_2030|M.thermoresistible__bu -------
MAB_4544c|M.abscessus_ATCC_199 -------
MLBr_01244|M.leprae_Br4923 AGWLSSS