For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRMTMSGALDAGLRSALEDDDKVIVMGEDVGKLGGVFRVTDGLQKDFGDHRVIDTPLAESGIIGTAVGLA MRGYRPVCEIQFDGFVYPAFDQIVSQVAKLHYRTKGAVGMPLTIRIPFGGGIGAVEHHSESPEAYFAHTA GLRVVSCSSPQDAYDMIRQAVACDDPVVFFEPKRRYWEKGEVDTALVPVPLGAARVVRSGTAATIAAYGP MVAVANAAAELAATEGISVEVVDLRSLSPVDFDTLEASVRKTGRLIVVHEAPVFMGLGAEIAARITERCF YHLEAPVLRVGGFALPYPANKVEHHYLPDAERVMDAVDRAIAA*
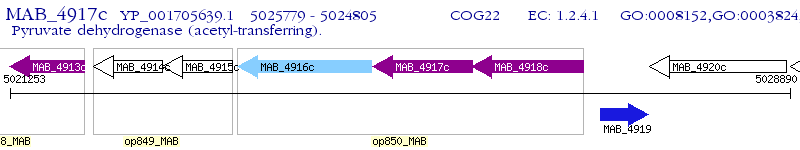
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4917c | - | - | 100% (324) | pyruvate dehydrogenase E1 component beta subunit |
| M. abscessus ATCC 19977 | MAB_0896c | - | e-106 | 59.06% (320) | putative pyruvate dehydrogenase E1 component, beta subunit |
| M. abscessus ATCC 19977 | MAB_2990c | - | 7e-10 | 29.03% (248) | 1-deoxy-D-xylulose-5-phosphate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2524c | pdhB | 5e-98 | 55.31% (320) | pyruvate dehydrogenase E1 component beta subunit PdhB |
| M. gilvum PYR-GCK | Mflv_3923 | - | 4e-07 | 27.46% (244) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. tuberculosis H37Rv | Rv2496c | pdhB | 1e-97 | 55.31% (320) | pyruvate dehydrogenase E1 component beta subunit PdhB |
| M. leprae Br4923 | MLBr_01038 | dxs | 2e-08 | 28.63% (255) | 1-deoxy-D-xylulose-5-phosphate synthase |
| M. marinum M | MMAR_3843 | pdhB | 1e-100 | 56.04% (323) | pyruvate dehydrogenase E1 component (beta subunit) PdhB |
| M. avium 104 | MAV_1676 | - | 1e-100 | 56.56% (320) | pyruvate dehydrogenase E1 component subunit beta |
| M. smegmatis MC2 155 | MSMEG_4711 | - | 1e-104 | 57.45% (322) | pyruvate dehydrogenase E1 component subunit beta |
| M. thermoresistible (build 8) | TH_0717 | pdhB | 1e-106 | 58.75% (320) | PROBABLE PYRUVATE DEHYDROGENASE E1 COMPONENT (BETA SUBUNIT) |
| M. ulcerans Agy99 | MUL_3774 | pdhB | 9e-99 | 55.11% (323) | pyruvate dehydrogenase E1 component (beta subunit) PdhB |
| M. vanbaalenii PYR-1 | Mvan_4085 | - | 1e-101 | 57.68% (319) | transketolase, central region |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4711|M.smegmatis_MC2_155 --MTQIIDRPAAQGDAPE---PWGSILTPAAAQAPVPAV----TELTMVQ
TH_0717|M.thermoresistible__bu MTVTQLIERPLVT------------------ARPPVP-------ELTLAQ
Mvan_4085|M.vanbaalenii_PYR-1 --MTQLIERPFGFDDDAA---E----PQRTPVAVPVAPI----APISMAQ
Mb2524c|M.bovis_AF2122/97 --MTQIADRPARPDE---------------TLAVAVSDI---TQSLTMVQ
Rv2496c|M.tuberculosis_H37Rv --MTQIADRPARPDE---------------TLAVAVSDI---TQSLTMVQ
MMAR_3843|M.marinum_M --MTQIADPPARPGP---------------APAAPVVGT---MQSLTMVQ
MUL_3774|M.ulcerans_Agy99 --MTQIADPPARPGP---------------APAAPVVGT---MQSLTMVQ
MAV_1676|M.avium_104 --MTQLADRPTGHGE---------------TLTAAVQDVALGAQALTMVQ
MAB_4917c|M.abscessus_ATCC_199 --MTR----------------------------------------MTMSG
Mflv_3923|M.gilvum_PYR-GCK --MLEQIRGPADLQHLSRAQMEDLAREIRDFLIHKVAATGGHLGPNLGVV
MLBr_01038|M.leprae_Br4923 --MLEQIRRPADLQHLSQQQLRDLAAEIRELLVHKVAATGGHLGPNLGVV
: .
MSMEG_4711|M.smegmatis_MC2_155 AINRALRDAMAADERVLVFG------------------------------
TH_0717|M.thermoresistible__bu AVNRALHDAMAADQRVLVFG------------------------------
Mvan_4085|M.vanbaalenii_PYR-1 AINRALHDAMRVDDRVLVFG------------------------------
Mb2524c|M.bovis_AF2122/97 AINRALYDAMAADERVLVFG------------------------------
Rv2496c|M.tuberculosis_H37Rv AINRALYDAMAADERVLVFG------------------------------
MMAR_3843|M.marinum_M ALNQALHDAMAADDRVLVFG------------------------------
MUL_3774|M.ulcerans_Agy99 ALNQALHDAMAADDRVLVFG------------------------------
MAV_1676|M.avium_104 ALNRALHDAMAADERVLVFG------------------------------
MAB_4917c|M.abscessus_ATCC_199 ALDAGLRSALEDDDKVIVMG------------------------------
Mflv_3923|M.gilvum_PYR-GCK ELTLALHRVFDSPHDPILFDTGHQAYVHKMLTGRSRDFDTLRKKDGLSGY
MLBr_01038|M.leprae_Br4923 ELTLALHRVFDSPHDPIIFDTGHQAYVHKMLTGRCQDFDSLRKKAGLSGY
: .* .: . :::.
MSMEG_4711|M.smegmatis_MC2_155 ----------EDVATLGGVFRVTDGLTESFGAE----RCFDTPLAESAII
TH_0717|M.thermoresistible__bu ----------QDVARLGGVFRVTDGLSAAFGEE----RCFDTPLAESAII
Mvan_4085|M.vanbaalenii_PYR-1 ----------EDVATLGGVFRVTEGLAETYGEQ----RCFDTPLAESAII
Mb2524c|M.bovis_AF2122/97 ----------EDVAVEGGVFRVTEGLADTFGAD----RCFDTPLAESAII
Rv2496c|M.tuberculosis_H37Rv ----------EDVAVEGGVFRVTEGLADTFGAD----RCFDTPLAESAII
MMAR_3843|M.marinum_M ----------EDVGIAGGVFRVTEGLAETFGEH----RCFDTPLAESALI
MUL_3774|M.ulcerans_Agy99 ----------EDVGIAGGVFRVTEGLAETFGEH----RCFDTPLAESALI
MAV_1676|M.avium_104 ----------EDVAVQGGVFRVTEGLAEAFGES----RCFDTPLAESAII
MAB_4917c|M.abscessus_ATCC_199 ----------EDVGKLGGVFRVTDGLQKDFGDH----RVIDTPLAESGII
Mflv_3923|M.gilvum_PYR-GCK PSRSESEHDWVESSHASSALSYADGLAKAFELNGHRNRHVVAVVGDGALT
MLBr_01038|M.leprae_Br4923 PSRAESEHDWVESSHASTALSYADGLAKAFELAGNRNRHVVAVVGDGALT
: . . .: ::** : * . : :.:..:
MSMEG_4711|M.smegmatis_MC2_155 G----IAVGFAIRGFVPVPEIQFDG--FSYPAFDQIASHLAKYRMRTHGD
TH_0717|M.thermoresistible__bu G----IAIGLAIRGFVPVPEIQFDG--FSYPAFDQIVSHLAKYRMRTRGD
Mvan_4085|M.vanbaalenii_PYR-1 G----IAVGMAIRGLVPVPEIQFDG--FAAPAFDQMVSHLAKYRMRTRGD
Mb2524c|M.bovis_AF2122/97 G----IAVGLALRGFVPVPEIQFDG--FSYPAFDQVVSHLAKYRTRTRGE
Rv2496c|M.tuberculosis_H37Rv G----IAVGLALRGFVPVPEIQFDG--FSYPAFDQVVSHLAKYRTRTRGE
MMAR_3843|M.marinum_M G----IAVGLALRGFVPVPEIQFDG--FSYPAFDQVVSHLAKYRTRTRGE
MUL_3774|M.ulcerans_Agy99 G----IAVGLALRGFVPVPEIQFDD--FSYPAFDQVVSHLAKYRTRTRGE
MAV_1676|M.avium_104 G----IAVGLALRGFVPVPEIQFDG--FSYPAFDQVVSHLAKYRTRTRGA
MAB_4917c|M.abscessus_ATCC_199 G----TAVGLAMRGYRPVCEIQFDG--FVYPAFDQIVSQVAKLHYRTKGA
Mflv_3923|M.gilvum_PYR-GCK GGMCWEALNNIAASRRPVVIVVNDNGRSYAPTIGGFAEHLAGLRLQPGYE
MLBr_01038|M.leprae_Br4923 GGMCWEALNNIAATPRPVVIVVNDNGRSYAPTIGGVADHLATLRLQPAYE
* *:. ** : *. *::. ...::* : :.
MSMEG_4711|M.smegmatis_MC2_155 IDMP----VTVRIPSFGGIGAVEHHSEST-------ETYWLHTAGLKVVV
TH_0717|M.thermoresistible__bu VEMR----VTVRIPSFGGIGAVEHHSEST-------ETYWLHTAGLKVVA
Mvan_4085|M.vanbaalenii_PYR-1 VDMP----VTVRIPSFGGIGAVEHHSEST-------ETYWLHTAGLKVVT
Mb2524c|M.bovis_AF2122/97 VDMP----VTVRIPSFGGIGAAEHHSDST-------ESYWVHTAGLKVVV
Rv2496c|M.tuberculosis_H37Rv VDMP----VTVRIPSFGGIGAAEHHSDST-------ESYWVHTAGLKVVV
MMAR_3843|M.marinum_M VNMA----VTVRIPSFGGIGAAEHHSDST-------ESYWAHTAGLKVVV
MUL_3774|M.ulcerans_Agy99 VNMA----VTVRIPSFGGIGAAEHHLDAT-------ESYWAHTAGLKVVV
MAV_1676|M.avium_104 IDMP----VTVRVPSFGGIGAAEHHSEST-------ESYWAHTAGLKVVV
MAB_4917c|M.abscessus_ATCC_199 VGMP----LTIRIPFGGGIGAVEHHSESP-------EAYFAHTAGLRVVS
Mflv_3923|M.gilvum_PYR-GCK RVLEEGRKAVRGVPMIGEFCYQCMHSIKAGIKDALSPQVMFTDLGLKYVG
MLBr_01038|M.leprae_Br4923 RLLEKGRDALHSLPLIGQIAYRFMHSVKAGIKDSLSPQLLFTDLGLKYVG
: :* * : * . **: *
MSMEG_4711|M.smegmatis_MC2_155 PST---PSDAYWLLRESISSPDPVIYLEPKRR--------------YWAR
TH_0717|M.thermoresistible__bu PGT---PADAYWLLRHAITCPDPVIYLEPKRR--------------YWAR
Mvan_4085|M.vanbaalenii_PYR-1 PST---PTDAYWLLRYAIASRDPVIFLEPKRR--------------YWAK
Mb2524c|M.bovis_AF2122/97 PST---PGDAYWLLRHAIACPDPVMYLEPKRR--------------YHSR
Rv2496c|M.tuberculosis_H37Rv PST---PGDAYWLLRHAIACPDPVMYLEPKRR--------------YHGR
MMAR_3843|M.marinum_M PST---PGDAYWLLRHAIACPDPVMYLEPKRR--------------YQVR
MUL_3774|M.ulcerans_Agy99 PST---PGDAYWLLRHAIACPDPVMYLEPKRR--------------YQVR
MAV_1676|M.avium_104 PSN---PADAYWLLRHAIACPDPVMYLEPKRR--------------YQGR
MAB_4917c|M.abscessus_ATCC_199 CSS---PQDAYDMIRQAVACDDPVVFFEPKRR--------------YWEK
Mflv_3923|M.gilvum_PYR-GCK PIDGHDESAVEGALRHARAFNAPVVVHVVTRKGMGYAPAENDVDEQMHAC
MLBr_01038|M.leprae_Br4923 PVDGHDEHAVEVALRKARGFGGPVIVHVVTRKGMGYPPAEADQAEQMHTC
. :* : **: .*:
MSMEG_4711|M.smegmatis_MC2_155 EAVDTTT---------------PALPIGRAAVRRNGSDVTVITYGGLVAT
TH_0717|M.thermoresistible__bu GPVDTTT---------------PAGPLGRAAVARSGTDVTVITYGSLVTT
Mvan_4085|M.vanbaalenii_PYR-1 EVVDTGN---------------PADPIGRAAIRRAGDDVTVLTYGPLVAT
Mb2524c|M.bovis_AF2122/97 GMVDTSR---------------PEPPIGHAMVRRSGTDVTVVTYGNLVST
Rv2496c|M.tuberculosis_H37Rv GMVDTSR---------------PEPPIGHAMVRRSGTDVTVVTYGNLVST
MMAR_3843|M.marinum_M GPVDTSR---------------PEPAIGQAMIRRAGADVTVITYGNLVST
MUL_3774|M.ulcerans_Agy99 GPVDTSR---------------PEPAIGQAMIRRAGADVTVITYGNLVST
MAV_1676|M.avium_104 GLVDAGR---------------PEPPIGRAMVRRAGTDVTVVTYGSLVAT
MAB_4917c|M.abscessus_ATCC_199 GEVDTAL---------------VPVPLGAARVVRSGTAATIAAYGPMVAV
Mflv_3923|M.gilvum_PYR-GCK GVIDPETGLATSVPGPGWTAAFSETLIELAAKRRDIVAITAAMPGPTGLS
MLBr_01038|M.leprae_Br4923 GVMDPTTGQPTKIAAPDWTAIFSDALIGYAMKRRDIVAITAAMPGPTGLT
:*. : * * * *
MSMEG_4711|M.smegmatis_MC2_155 AL-----------SAAELAED-RGWSMEVVDLRSLNPLDFDTVADSVRRT
TH_0717|M.thermoresistible__bu AL-----------SAAELAADTHGWSLEVVDLRSLNPLDFDTIAESVRRT
Mvan_4085|M.vanbaalenii_PYR-1 AL-----------NAAELSPH----GLEVVDLRSLNPLDFDTVAASVRKT
Mb2524c|M.bovis_AF2122/97 AL-----------SSADTAEQQHDWSLEVIDLRSLAPLDFDTIAASIQRT
Rv2496c|M.tuberculosis_H37Rv AL-----------SSADTAEQQHDWSLEVIDLRSLAPLDFDTIAASIQRT
MMAR_3843|M.marinum_M AL-----------SAAEDAAHQQGWSLEVIDLRSLIPLDFETIAASIRRT
MUL_3774|M.ulcerans_Agy99 AL-----------SAAEDAAHQQGWSLEVIDLRSLIPLDFETIAASIRRT
MAV_1676|M.avium_104 AV-----------GAAEEAQHQRGWSLEVIDLRSLVPLDFDTIATSIHRT
MAB_4917c|M.abscessus_ATCC_199 AN-----------AAAELAAT-EGISVEVVDLRSLSPVDFDTLEASVRKT
Mflv_3923|M.gilvum_PYR-GCK AFRKRFPDRFFDVGIAEQHAMTSAAGLAMGGMHPVVALYSTFLNRAFDQM
MLBr_01038|M.leprae_Br4923 AFGQCFPDRLFDVGIAEQHAMTSAAGLAMGRMHPVVAIYSTFLNRAFDQI
* *: .: : ::.: .: : :. :
MSMEG_4711|M.smegmatis_MC2_155 GRAVVMHEGPRTLGFG-------------------------------AEL
TH_0717|M.thermoresistible__bu GRAVVLHEGPRTLGFG-------------------------------AEL
Mvan_4085|M.vanbaalenii_PYR-1 GRAVVMHEGARTVGFG-------------------------------AEL
Mb2524c|M.bovis_AF2122/97 GRCVVMHEGPRSLGYG-------------------------------AGL
Rv2496c|M.tuberculosis_H37Rv GRCVVMHEGPRSLGYG-------------------------------AGL
MMAR_3843|M.marinum_M GRCVVLHEGPRSLGYG-------------------------------AGL
MUL_3774|M.ulcerans_Agy99 GRCVVLHEGPRSLGYG-------------------------------AGL
MAV_1676|M.avium_104 GRCVVMHEGPRTLGYG-------------------------------AEL
MAB_4917c|M.abscessus_ATCC_199 GRLIVVHEAPVFMGLG-------------------------------AEI
Mflv_3923|M.gilvum_PYR-GCK LMDVALHKLPVTMVLDRSGITGPDGASHNGMWDLSILGIVPGMRVAAPRD
MLBr_01038|M.leprae_Br4923 MMDVALHKLPVTMVIDRAGITGSDGPSHNGMWDLSMLGIVPGMRVAAPRD
:.:*: . : . .
MSMEG_4711|M.smegmatis_MC2_155 AARISEELFYDLEAP-----------------------------------
TH_0717|M.thermoresistible__bu AARITEELFYDLEAP-----------------------------------
Mvan_4085|M.vanbaalenii_PYR-1 AARISEECFYDLEAP-----------------------------------
Mb2524c|M.bovis_AF2122/97 AARIQEEMFYQLEAP-----------------------------------
Rv2496c|M.tuberculosis_H37Rv AARIQEEMFYQLEAP-----------------------------------
MMAR_3843|M.marinum_M AARIQEELFYELEAP-----------------------------------
MUL_3774|M.ulcerans_Agy99 AARIQEELFYELEAP-----------------------------------
MAV_1676|M.avium_104 AARIQEELFYELEAP-----------------------------------
MAB_4917c|M.abscessus_ATCC_199 AARITERCFYHLEAP-----------------------------------
Mflv_3923|M.gilvum_PYR-GCK GARLREELAEAVEVKDGPTAVRFPKGDVGEDIPAIERRDGVDVLAVPADG
MLBr_01038|M.leprae_Br4923 AIRLREELGEALDVDDGPTAIRFPKGDVCEDIPALKRRSGVDVLAVPATG
. *: *. ::.
MSMEG_4711|M.smegmatis_MC2_155 --------------------------------------------------
TH_0717|M.thermoresistible__bu --------------------------------------------------
Mvan_4085|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2524c|M.bovis_AF2122/97 --------------------------------------------------
Rv2496c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3843|M.marinum_M --------------------------------------------------
MUL_3774|M.ulcerans_Agy99 --------------------------------------------------
MAV_1676|M.avium_104 --------------------------------------------------
MAB_4917c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3923|M.gilvum_PYR-GCK LSDDVLIIAVGAFAAMSVAVAERLRDQGIGVTVVDPRWVLPVPAAINTLA
MLBr_01038|M.leprae_Br4923 LAQDVLLVGVGVFASMALAVAKRLHNQGIGVTVIDPRWVLPVCDGVLELA
MSMEG_4711|M.smegmatis_MC2_155 ------------------------VLRATGFDTPYPPARLEKLWLPGVDR
TH_0717|M.thermoresistible__bu ------------------------VLRATGFDTPYPPARLEKLWLPGPDR
Mvan_4085|M.vanbaalenii_PYR-1 ------------------------VLRATGFDTPYPPARLEKLWLPGVDR
Mb2524c|M.bovis_AF2122/97 ------------------------VLRACGFDTPYPPARLEKLWLPGPDR
Rv2496c|M.tuberculosis_H37Rv ------------------------VLRACGFDTPYPPARLEKLWLPGPDR
MMAR_3843|M.marinum_M ------------------------VLRACGFDTPYPPARLERLWLPGPDR
MUL_3774|M.ulcerans_Agy99 ------------------------VLRACGFDTPYPPARLERLWLPGPDR
MAV_1676|M.avium_104 ------------------------VLRACGFDTPYPPARLEKWWLPGPDR
MAB_4917c|M.abscessus_ATCC_199 ------------------------VLRVGGFALPYPANKVEHHYLPDAER
Mflv_3923|M.gilvum_PYR-GCK AAHKLVVTVEDNGGHGGVGSAVSAALRRAEIDVPCRDAALPQAFFDHASR
MLBr_01038|M.leprae_Br4923 HTHKLIVTLEDNGVNGGVGAAVSTALRQVEIDTPCRDVGLPQEFYDHASR
.** : * : : : .*
MSMEG_4711|M.smegmatis_MC2_155 LLDCVERAMEQP---------------------------------
TH_0717|M.thermoresistible__bu LLDCVERSLGYP---------------------------------
Mvan_4085|M.vanbaalenii_PYR-1 LLDCVDKAMGQP---------------------------------
Mb2524c|M.bovis_AF2122/97 LLDCVERVLRQP---------------------------------
Rv2496c|M.tuberculosis_H37Rv LLDCVERVLRQP---------------------------------
MMAR_3843|M.marinum_M LLDCVERVLGQP---------------------------------
MUL_3774|M.ulcerans_Agy99 LLDCVERVLGQP---------------------------------
MAV_1676|M.avium_104 LLDCVERTLELP---------------------------------
MAB_4917c|M.abscessus_ATCC_199 VMDAVDRAIAA----------------------------------
Mflv_3923|M.gilvum_PYR-GCK GEVLAEVGLTERTIARQITGWVAALG-----ATAGDRQVSESVD-
MLBr_01038|M.leprae_Br4923 SEVLADLGLTDQDVARRITGWVVAFGHCGSGDDAGQYGPRSSQTM
.: :