For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRELTDIEMLSELEPIAETNLNRHLAVATEWHPHDYVPWDRGRNFAQMGGDDWDPEQSQLSEVARAAMIT NLLTEDNLPSYHRQAAKYFGLDGAWGTWVGRWTAEENRHGIVIRDYLVVTRGVDPVALERARMEHMTAGF DATDEEESVHQTDFLLSVAYTTLQELATRVSHRNTGKVCDDPVADRMLQRVAADENLHMIFYRNMCSAAL DLVPDQALDAIGAVIENFRMPGQGMPNFRRNGVLMAKHGIYDPRQHLDEVVTPNLRKWRIFERNDFSAKG EARRDQLAAYVEDLKGQVVKFEEQRDRMLAREARKREARAG*
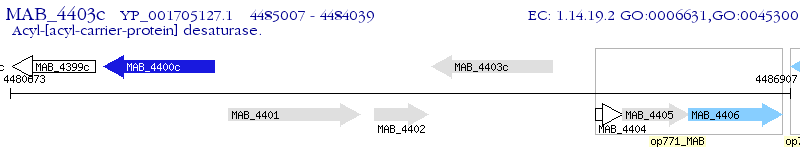
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4403c | - | - | 100% (322) | acyl- |
| M. abscessus ATCC 19977 | MAB_2550c | - | e-115 | 59.63% (322) | fatty acid desaturase |
| M. abscessus ATCC 19977 | MAB_0754c | - | e-114 | 59.01% (322) | fatty acid desaturase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0847c | desA1 | 1e-108 | 56.33% (316) | acyl- |
| M. gilvum PYR-GCK | Mflv_1668 | - | 1e-109 | 56.17% (324) | fatty acid desaturase, type 2 |
| M. tuberculosis H37Rv | Rv0824c | desA1 | 1e-108 | 56.33% (316) | acyl- |
| M. leprae Br4923 | MLBr_02185 | desA1 | 1e-105 | 54.23% (319) | acyl- |
| M. marinum M | MMAR_4856 | desA1 | 1e-109 | 56.56% (320) | acyl- |
| M. avium 104 | MAV_1793 | - | 1e-105 | 55.17% (319) | Fatty acid desaturase |
| M. smegmatis MC2 155 | MSMEG_5773 | - | 1e-110 | 57.23% (318) | fatty acid desaturase |
| M. thermoresistible (build 8) | TH_1577 | - | 1e-107 | 55.28% (322) | fatty acid desaturase |
| M. ulcerans Agy99 | MUL_0445 | desA1 | 1e-107 | 55.94% (320) | acyl- |
| M. vanbaalenii PYR-1 | Mvan_5085 | - | 1e-108 | 55.25% (324) | fatty acid desaturase, type 2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0847c|M.bovis_AF2122/97 MS-AKLTDLQLLHELEPVVEKYLNRHLSMHKPWNPHDYIPWSDGKNYYAL
Rv0824c|M.tuberculosis_H37Rv MS-AKLTDLQLLHELEPVVEKYLNRHLSMHKPWNPHDYIPWSDGKNYYAL
MMAR_4856|M.marinum_M MS-APLTDLHLLHELEPVVEKYLNRHLSMHKNWNPHDYIPWSDGKNYYAL
MUL_0445|M.ulcerans_Agy99 MS-APLTDLHLLHELEPVVEKYLNRHLSIHKNWNPHDYIPWSDGKNYYAL
MLBr_02185|M.leprae_Br4923 MS-AELTDLQLLHELEPVVEKYMNRHERLHKNWNPHDYIPWSDGKNFYAL
MAV_1793|M.avium_104 MP-KDLTNLQLLHELEPVVEGLLNRHLAKFKEWNPHDYVPWSDGKNFHTR
Mflv_1668|M.gilvum_PYR-GCK MS-KDLTDLQLLTELEPVVEPLVNRHMRMKKDWNPHDYIPWSDGKNYYAL
Mvan_5085|M.vanbaalenii_PYR-1 MS-KDLTDLQLLTELEPVVEPLINRHMRMTKDWNPHDYIPWSDGKNYYAL
MSMEG_5773|M.smegmatis_MC2_155 MSTDDYTDLQLLHELEPVVETLIDRHMRMCKDWNPHDYIPWSDGKNYYAL
TH_1577|M.thermoresistible__bu MH-ENLTDLHLLRELEPVVEKLLNRHLSMSKDWNPHDYIPWSDGKNYYAL
MAB_4403c|M.abscessus_ATCC_199 MP-RELTDIEMLSELEPIAETNLNRHLAVATEWHPHDYVPWDRGRNFAQM
* *::.:* ****:.* ::** . *:****:**. *:*:
Mb0847c|M.bovis_AF2122/97 GGQDWDPDQSKLSDVAQVAMVQNLVTEDNLPSYHREIAMNMGMDGAWGQW
Rv0824c|M.tuberculosis_H37Rv GGQDWDPDQSKLSDVAQVAMVQNLVTEDNLPSYHREIAMNMGMDGAWGQW
MMAR_4856|M.marinum_M GGQEWDPEQSKLSDVAQIAMVQNLVTEDNLPSYHREIAMNMGMDGAWGQW
MUL_0445|M.ulcerans_Agy99 GGQEWDPEQSKLSDVAQIAMVQNLVTEDNLPSYHREIAMNMGMDGAWGQW
MLBr_02185|M.leprae_Br4923 GGQDWDPEQSQLSDVAQVAMVQNLVTEDNLPSYHREIAMTMGMDGAWGQW
MAV_1793|M.avium_104 GGQDWEPGQTRLPDVAQVAIVQNLLTEDNLPSYHREIAMNFGMDGPWGQW
Mflv_1668|M.gilvum_PYR-GCK GGQDWDPEQSQLSDVARVAMLVNLLTEDNLPSYHREIAMNFSMDGPWGFW
Mvan_5085|M.vanbaalenii_PYR-1 GGQDWDPEQAKLSEVARVAMMVNLLTEDNLPSYHREIAMNFTMDGPWGFW
MSMEG_5773|M.smegmatis_MC2_155 GGQDWDPEQSKLSEVARTAMVQNLLTEDNLPSYHREIAMNMGMDGAWGQW
TH_1577|M.thermoresistible__bu GGQDWYPEEAKLSEVAQTAMVQNLLTEDNLPAYHREIAMNLGMDGAWGQW
MAB_4403c|M.abscessus_ATCC_199 GGDDWDPEQSQLSEVARAAMITNLLTEDNLPSYHRQAAKYFGLDGAWGTW
**::* * :::*.:**: *:: **:******:***: * : :**.** *
Mb0847c|M.bovis_AF2122/97 VNRWTAEENRHGIALRDYLVVTRSVDPVELEKLRLEVVNRGFSPGQNH--
Rv0824c|M.tuberculosis_H37Rv VNRWTAEENRHGIALRDYLVVTRSVDPVELEKLRLEVVNRGFSPGQNH--
MMAR_4856|M.marinum_M VNRWTAEENRHGIALRDYLVVTRSVDPVELEKLRLEVVNRGFSPGQNH--
MUL_0445|M.ulcerans_Agy99 VNRWTAEENRHSIALRDYLVVTRSVDPVELEKLRLEVVNRGFSPGQNH--
MLBr_02185|M.leprae_Br4923 VNRWTAEENRHGIALRDYLVVTRAVDPVELEKLRIEVVNRGFSPGQNH--
MAV_1793|M.avium_104 VNRWTAEEARHSTALRDYLVVTRSVDPVELELLRLEQMTRGFSPGQNQ--
Mflv_1668|M.gilvum_PYR-GCK VNRWTAEENRHGIALRDYLVVTRNVDPVELEMLRVEQMTRGFSPGQNDQI
Mvan_5085|M.vanbaalenii_PYR-1 VNRWTAEENRHGIALRDYLVVTRNVDPVELEMLRLEQMTRGFSPGQNEQI
MSMEG_5773|M.smegmatis_MC2_155 VNRWTAEENRHGIALRDYLVVTRACDPVELEELRVEQVTRGFSPGQNQ--
TH_1577|M.thermoresistible__bu VNRWTAEENRHGISLRDYLVVTRACDPVELEELRVEQMTRGFSPGQNQ--
MAB_4403c|M.abscessus_ATCC_199 VGRWTAEENRHGIVIRDYLVVTRGVDPVALERARMEHMTAGFDATDEE--
*.****** **. :******** *** ** *:* :. **.. ::.
Mb0847c|M.bovis_AF2122/97 QGHYFAESLTDSVLYVSFQELATRISHRNTGKACNDPVADQLMAKISADE
Rv0824c|M.tuberculosis_H37Rv QGHYFAESLTDSVLYVSFQELATRISHRNTGKACNDPVADQLMAKISADE
MMAR_4856|M.marinum_M QGDYFAESVTDSVIYVSFQELATRVSHRNTGKACNDPIADQLMAKISADE
MUL_0445|M.ulcerans_Agy99 QGDYFAESVTDSVIYVSFQELATRVSHRNTGKACNDLIADQLMAKISADE
MLBr_02185|M.leprae_Br4923 QGGLFADTLFDSILYVTFQELATRVSHRNTGKACNETIADQLLAKISSDE
MAV_1793|M.avium_104 QGDMFAESLFDSVMYVSFQELATRVSHRNTGKVCNDAIAEQLMARVSHDE
Mflv_1668|M.gilvum_PYR-GCK EADLFADSLFDSVIYVTFQELATRVSHRNTGKACEEPIADQLLQRVSADE
Mvan_5085|M.vanbaalenii_PYR-1 EADLFADSLFDSVIYVTFQELATRVSHRNTGKACEEPVADQLLQRVSADE
MSMEG_5773|M.smegmatis_MC2_155 QGDLFAESLFDSVMYVSFQELATRVSHRNTGKACNDPVADQLLQRVSADE
TH_1577|M.thermoresistible__bu QGDLFAESLFDSVIYVTFQELATRVSHRNTGKACNETVADQLLARISADE
MAB_4403c|M.abscessus_ATCC_199 -ESVHQTDFLLSVAYTTLQELATRVSHRNTGKVCDDPVADRMLQRVAADE
. . *: *.::******:*******.*:: :*:::: ::: **
Mb0847c|M.bovis_AF2122/97 NLHMIFYRDVSEAAFDLVPNQAMKSLHLILSHFQMPGFQVPEFRRKAVVI
Rv0824c|M.tuberculosis_H37Rv NLHMIFYRDVSEAAFDLVPNQAMKSLHLILSHFQMPGFQVPEFRRKAVVI
MMAR_4856|M.marinum_M NLHMIFYRDVSEAAFDLAPNQAMKSLHLILHNFKMPGFQVPEFRRKAVII
MUL_0445|M.ulcerans_Agy99 NLHMIFYRDVSEAAFDLAPNQAMKSLHLILHNFKMPGFQVPEFRRKAVII
MLBr_02185|M.leprae_Br4923 NLHMIFYRDVSEAGIELAPNLAIKALHKILHNFKMPGFLVPEFRRKAVII
MAV_1793|M.avium_104 NLHMIFYRDVSAAGLDIAPNQAMKSVHRILRNFKMPGFTVPEFRRKAVII
Mflv_1668|M.gilvum_PYR-GCK NLHMIFYRDVSEAGFEIAPDQAMRSLHKVLLNFKMPGYTIPDFRRRAVTI
Mvan_5085|M.vanbaalenii_PYR-1 NLHMIFYRDVSEAGFEIAPDQAMKSLHKVLRNFKMPGYTIPDFRRRAVTI
MSMEG_5773|M.smegmatis_MC2_155 NLHMIFYRDVSAAGLDIAPNQAMASLHRVLANFKMPGYTVPDFRRKAVVI
TH_1577|M.thermoresistible__bu NLHMIFYRDVSEAGFEIAPDQAMHSLHRVLRNFKMPGFTVPEFRRKAVVI
MAB_4403c|M.abscessus_ATCC_199 NLHMIFYRNMCSAALDLVPDQALDAIGAVIENFRMPGQGMPNFRRNGVLM
********::. *.:::.*: *: :: :: :*:*** :*:***..* :
Mb0847c|M.bovis_AF2122/97 AVGGVYDPRIHLDEVVMPVLKKWRIFEREDFTGEGAKLRDELALVIKDLE
Rv0824c|M.tuberculosis_H37Rv AVGGVYDPRIHLDEVVMPVLKKWRIFEREDFTGEGAKLRDELALVIKDLE
MMAR_4856|M.marinum_M AVGGVYDPRIHLDEVVMPVLKKWRIFEREDFTGEGAKMRDELALLVKELE
MUL_0445|M.ulcerans_Agy99 AVGGVYDPRIHLDEVVMPVLKKWRIFEREDFTGEGAKMRDELALLVKELE
MLBr_02185|M.leprae_Br4923 AVGGVYDIRIHLDEVVMPVLKKWRILEREDFSGEGTRLRDEIGEHLTELE
MAV_1793|M.avium_104 AVGGVYDPRIHLDEVVMPVLKKWRIFEREDFTGEAARMRDDLGLLVKELQ
Mflv_1668|M.gilvum_PYR-GCK ASGGVYDPRIHLEDIVMPVLKKWRIFEREDFNGESARMRDEIGAHVEELQ
Mvan_5085|M.vanbaalenii_PYR-1 ASGGVYDPRIHLEDIVMPVLKKWRIFEREDFNGESARLRDDLGLLVEELQ
MSMEG_5773|M.smegmatis_MC2_155 AVGGVYDPRIHLDDVVMPVLRKWRIFEREDFTGEGARLRDDLGRLVEELE
TH_1577|M.thermoresistible__bu AVGGVYDPRIHLDDVVMPVLKKWRIFEREDFTGEAARMRDDLGKLVEELE
MAB_4403c|M.abscessus_ATCC_199 AKHGIYDPRQHLDEVVTPNLRKWRIFERNDFSAKGEARRDQLAAYVEDLK
* *:** * **:::* * *:****:**:**..:. **::. : :*:
Mb0847c|M.bovis_AF2122/97 LACDKFEVSKQRQLDREARTGKKVSAHELHKTAGKLAMSRR
Rv0824c|M.tuberculosis_H37Rv LACDKFEVSKQRQLDREARTGKKVSAHELHKTAGKLAMSRR
MMAR_4856|M.marinum_M MSCDKFEESKQRQLDREARTGKRVSAFELHKTAGKLPMSRR
MUL_0445|M.ulcerans_Agy99 MSCDKFEESKQRQLDREARTGKRVSAFELHKTAGKLPMSRR
MLBr_02185|M.leprae_Br4923 AACDKFEVSKQRYIEREARRTEKITARKVLSTEGTLRMSGR
MAV_1793|M.avium_104 DASEKFEESKQRYLEREARRTEKITASKVLQTKGTLTLSRH
Mflv_1668|M.gilvum_PYR-GCK ETVQKFEIAKQRREERLRQMAEKKAAKN-LLVGSSAS----
Mvan_5085|M.vanbaalenii_PYR-1 EAVDKFEVAKQRREERLRQMAEKKAAKN-LLVGSSAS----
MSMEG_5773|M.smegmatis_MC2_155 DACDKFEVAKERRLERERKMAEKREMKN-LLVSSSSS----
TH_1577|M.thermoresistible__bu EACEKFEVSKQRRLEREARQAEKAAEKKKSFAASTAS----
MAB_4403c|M.abscessus_ATCC_199 GQVVKFEEQRDRMLAREARKREARAG---------------
*** ::* * : :