For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARRDGDSWEITESVGTTALGVASARDAETRSPNPLISDPYAGHFLAAAGDGAWNAYRFDGEPPAALVEAE PRLVERIDAMRSYVACRTKFFDDFFGDASTSGIRQAVILASGLDARAWRLDWPRDAVLFELDLPRVLAFK AETLDKQGATPRIRHVPVAVDLRDDWPAALRAGGFDVGRPAAWIAEGLLPYLPPAAQDLLFERIDTLSAP GSRIAVENFGEDFFNPETLARQRERGQAFRRVAEEMQGQDIPDVADLWYLEERTDAGEFLAGRGWRVTSE TAVALLGRHGRDIPADLPDATPNSAFLFAQKAD*
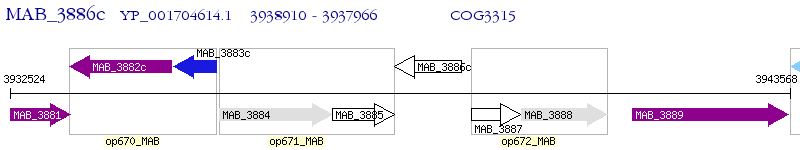
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3886c | - | - | 100% (314) | hypothetical protein MAB_3886c |
| M. abscessus ATCC 19977 | MAB_4587c | - | 3e-67 | 45.57% (316) | hypothetical protein MAB_4587c |
| M. abscessus ATCC 19977 | MAB_4586c | - | 4e-67 | 46.05% (304) | hypothetical protein MAB_4586c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3793c | - | 6e-84 | 51.45% (311) | hypothetical protein Mb3793c |
| M. gilvum PYR-GCK | Mflv_5025 | - | 2e-61 | 44.74% (304) | putative methyltransferase |
| M. tuberculosis H37Rv | Rv3767c | - | 6e-84 | 51.45% (311) | hypothetical protein Rv3767c |
| M. leprae Br4923 | MLBr_02640 | - | 3e-61 | 45.57% (305) | hypothetical protein MLBr_02640 |
| M. marinum M | MMAR_0955 | - | 3e-87 | 53.67% (313) | hypothetical protein MMAR_0955 |
| M. avium 104 | MAV_4558 | - | 5e-92 | 56.23% (313) | methyltransferase, putative, family protein |
| M. smegmatis MC2 155 | MSMEG_1480 | - | 5e-68 | 46.13% (310) | methyltransferase |
| M. thermoresistible (build 8) | TH_2759 | - | 8e-67 | 49.36% (312) | PUTATIVE methyltransferase |
| M. ulcerans Agy99 | MUL_0706 | - | 2e-86 | 53.35% (313) | hypothetical protein MUL_0706 |
| M. vanbaalenii PYR-1 | Mvan_1344 | - | 3e-66 | 46.79% (312) | putative methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_02640|M.leprae_Br4923 ------------MRTHDDTWDIKTSVGTTAVMVAAARAAETDRPDALIRD
TH_2759|M.thermoresistible__bu MVMMFDVTEAASARSEGDSWDITESVGATALGVAAERALETSQPDPLIRD
Mflv_5025|M.gilvum_PYR-GCK -----------MARTPDDSWDLASSVGATATMVAAGRAVASADPDPLIND
Mvan_1344|M.vanbaalenii_PYR-1 -----------MARTADDSWDPASSVGATATMVAAGRAAASADPDPLIND
MSMEG_1480|M.smegmatis_MC2_155 -----------MTRTDGDTWDLATSVGSTATGVAAMRALATRQPDPLIDD
Mb3793c|M.bovis_AF2122/97 -----------MPRTDNDSWAITESVGATALGVAAARAAETESDNPLIND
Rv3767c|M.tuberculosis_H37Rv -----------MPRTDNDSWAITESVGATALGVAAARAAETESDNPLIND
MMAR_0955|M.marinum_M -----------MARTEDDSWEITESVGATALGVASARAAETRSQHPLISD
MUL_0706|M.ulcerans_Agy99 -----------MARTEDDSWEITESVGATALGVASARAAETRSQHPLISD
MAV_4558|M.avium_104 -----------MPRTDDDSWEITESVGATALGVAAARAAETESENPLISD
MAB_3886c|M.abscessus_ATCC_199 -----------MARRDGDSWEITESVGTTALGVASARDAETRSPNPLISD
* .*:* ***:** **: * : ..** *
MLBr_02640|M.leprae_Br4923 PYAKLLVTNTGAGALWEAMLDPSMVAKVEAIDAEAAAMVEHMRSYQAVRT
TH_2759|M.thermoresistible__bu EYAFLLVS--AAGPPWTQLASSDLSWLGD--DETARRMHLMSRDYQAVRT
Mflv_5025|M.gilvum_PYR-GCK PYAEPLVR--AVGLDFFTRMLDGELDLSVFPDSSPER-AQAMIDGMAVRT
Mvan_1344|M.vanbaalenii_PYR-1 PYAEPLVR--AVGLSFFTKMLDGELDLSQFADGSPER-VQAMIDGMAVRT
MSMEG_1480|M.smegmatis_MC2_155 PYADALVK--AVGLEHCIALADGET----CVEGDPMLDLNRMCEQIAVRT
Mb3793c|M.bovis_AF2122/97 PFARIFVDAAGDGIWSMYTNR-TLLAGATDLDPDLRAPIQQMIDFMAART
Rv3767c|M.tuberculosis_H37Rv PFARIFVDAAGDGIWSMYTNR-TLLAGATDLDPDLRAPIQQMIDFMAART
MMAR_0955|M.marinum_M PFAQVFLDAVGDGVWNWHSAP-QLPAELLEIEPDLPLQMEAMVSYMASRT
MUL_0706|M.ulcerans_Agy99 PFAQVFLDAVGDGVWNWHSAP-QLPAELLEIEPDLPLQMEAMVSYMASRT
MAV_4558|M.avium_104 PFARVFLDAAGDGMWNWFAAP-NLPAQIAEAEPDLKPRMQGMVDYMAART
MAB_3886c|M.abscessus_ATCC_199 PYAGHFLAAAGDGAWNAYRFDGEPPAALVEAEPRLVERIDAMRSYVACRT
:* :: . * : . * **
MLBr_02640|M.leprae_Br4923 NFFDTYFNNAVIDGIRQFVILASGLDSRAYRLDWPTGTTVYEIDQPKVLA
TH_2759|M.thermoresistible__bu HYFDRYFAAAARAGIRQIVLLAAGLDSRAYRLDWPAGTVVYELDQPRVLD
Mflv_5025|M.gilvum_PYR-GCK RFFDDCCLAAASAGVRQVVILAAGLDARTYRLPWPDGTVVYELDQPDVIA
Mvan_1344|M.vanbaalenii_PYR-1 RFFDDCCGTSTTAGITQVVILASGLDARAYRLGWPAGTVVYELDQPAVIE
MSMEG_1480|M.smegmatis_MC2_155 RYFDELFIAAGADGVRQAVILASGLDTRAYRLDWPAGTVVFEVDQPQVIE
Mb3793c|M.bovis_AF2122/97 AFFDEYFLATADAGVRQVVILASGLDSRAWRLPWPDGTVVYELDQPKVLE
Rv3767c|M.tuberculosis_H37Rv AFFDEYFLATADAGVRQVVILASGLDSRAWRLPWPDGTVVYELDQPKVLE
MMAR_0955|M.marinum_M AFFDEFFLDATRAGIGQAVILAAGLDSRAWRLPWPAGTTVFELDQPRVLE
MUL_0706|M.ulcerans_Agy99 AFFDEFFLDATRAGIGQAVILAAGLDSRAWRLPWPAGTTVFELDQPRVLE
MAV_4558|M.avium_104 AFFDNFFLAATRAGVRQVVILAAGLDSRAWRLPFEDGTTVYELDQPRVLE
MAB_3886c|M.abscessus_ATCC_199 KFFDDFFGDASTSGIRQAVILASGLDARAWRLDWPRDAVLFELDLPRVLA
:** : *: * *:**:***:*::** : .:.::*:* * *:
MLBr_02640|M.leprae_Br4923 YKSTTLAEHGVTPTADRREVPIDLRQDWPPALRSAGFDPSARTAWLAEGL
TH_2759|M.thermoresistible__bu FKNRTLQAHGAVAKADRRTVAVDLRDDWPAALRHAGFDSAAPTAWLAEGL
Mflv_5025|M.gilvum_PYR-GCK FKTQTLRNLGAEPAATQRPVPVDLREDWPAALRAAGFDATRPTAWLAEGL
Mvan_1344|M.vanbaalenii_PYR-1 FKTRTLAGLGAHPTATHRPVPIDLREDWPAALHAAGFDASRPTAWLAEGL
MSMEG_1480|M.smegmatis_MC2_155 FKTRTLADLGAQPTAERRTVAVDLRDDWPAALRDAGFDPAEPTAWIAEGL
Mb3793c|M.bovis_AF2122/97 FKSATLRQHGAQPASQLVNVPIDLRQDWPKALQKAGFDPSKPCAWLAEGL
Rv3767c|M.tuberculosis_H37Rv FKSATLRQHGAQPASQLVNVPIDLRQDWPKALQKAGFDPSKPCAWLAEGL
MMAR_0955|M.marinum_M FKAATLAEHGAEPACGRVAVAVDLRQDWPTALRQAGFDPSVPSVWSAEGL
MUL_0706|M.ulcerans_Agy99 FKAATLAEHGAEPACGRVAVAVDLRQDWPTALRQAGFDPSVSSVWSAEGL
MAV_4558|M.avium_104 FKATTLAEYGARPTCHLVSVPVDLRHDWPAALRQAGFDAHAPSAWSAEGL
MAB_3886c|M.abscessus_ATCC_199 FKAETLDKQGATPRIRHVPVAVDLRDDWPAALRAGGFDVGRPAAWIAEGL
:* ** *. . *.:***.*** **: .*** .* ****
MLBr_02640|M.leprae_Br4923 LMYLPATAQDGLFTEIGGLSAVGSRIAVET---SPLHGDEWREQMQLRFR
TH_2759|M.thermoresistible__bu LCYLPADAQDRLFERITALSAPGSWLAVEAFHHDPQRATE-RRRAEWRDR
Mflv_5025|M.gilvum_PYR-GCK LIYLPPEAQDALFDTVTALSAPGSTLATEYVPGIVDFDGAKARALSEPLR
Mvan_1344|M.vanbaalenii_PYR-1 LIYLPPEAQDQLFDRITALSAPGSTIATEYAPGIIDFDDEKARAMSAPMR
MSMEG_1480|M.smegmatis_MC2_155 LIYLPAEAQDRLLDNITALSAPGSRLATEHMDHRILAEGAGKR-IGEWSR
Mb3793c|M.bovis_AF2122/97 VRYLPARAQDLLFERIDALSRPGSWLASNVPGAGFLDPERMRRQRADMRR
Rv3767c|M.tuberculosis_H37Rv VRYLPARAQDLLFERIDALSRPGSWLASNVPGAGFLDPERMRRQRADMRR
MMAR_0955|M.marinum_M MPYLPAVAQDLLFERVQGLTVRASRIAVEALGPKFLDPQVRANRSARMER
MUL_0706|M.ulcerans_Agy99 MPYLPAVAQDLLFERVQGLTVRASRIAVEALGPKFLDPQVRANRSARIER
MAV_4558|M.avium_104 LPFLPAAAQQLLFERVQTLAAPGSRIAVEAPGPDFLDEAARERQRQTMQR
MAB_3886c|M.abscessus_ATCC_199 LPYLPPAAQDLLFERIDTLSAPGSRIAVENFGEDFFNPETLARQRERGQA
: :**. **: *: : *: .* :* : .
MLBr_02640|M.leprae_Br4923 RVSDALGFEQAVDVQE---LIYHDENRAVVADWLNRHGWRATAQSAPDEM
TH_2759|M.thermoresistible__bu TARMKAALDLNLDFEA---LTYDDPDRADAGEWLAAHGWHPDSVPGTEEM
Mflv_5025|M.gilvum_PYR-GCK EHG------LDIDMPS---LVYTGA-RTPVMDHLRGAGWQVSGTARAELF
Mvan_1344|M.vanbaalenii_PYR-1 EMG------LDIDMPS---LVYAGT-RSHVMDYLAGQGWEVTGVPRRELF
MSMEG_1480|M.smegmatis_MC2_155 RVG------SSLDIAD---LFYTGE-RNTAGDYLRGLGWQVTVRPSKEAY
Mb3793c|M.bovis_AF2122/97 MRAAAAKLVET-EISDVDDLWYAEQ-RTAVAEWLRERGWDVSTATLPELL
Rv3767c|M.tuberculosis_H37Rv MRAAAAKLVET-EISDVDDLWYAEQ-RTAVAEWLRERGWDVSTATLPELL
MMAR_0955|M.marinum_M IRAVMAHVEPQREIPRTDELWYFEE-REDVGDWFRRHDWDVTVTPSGELM
MUL_0706|M.ulcerans_Agy99 IRAVMAHVEPQREIPRTDELWYFEE-REDVGDWFRRHDWDVTVTPSGELM
MAV_4558|M.avium_104 VRDLMADLEPDRDIPDVQDLWYFEE-REDVGDWLGRHGWDVTVTPAPELM
MAB_3886c|M.abscessus_ATCC_199 FRRVAEEMQGQ-DIPDVADLWYLEE-RTDAGEFLAGRGWRVTSETAVALL
:. * * * . : : .* .
MLBr_02640|M.leprae_Br4923 RRVGR--WGDGVPMADDKDAFAEFVTAHRL---
TH_2759|M.thermoresistible__bu ARMGRPVPADLLDVAVPSTLLRAHLNGASR---
Mflv_5025|M.gilvum_PYR-GCK ARFGRPLPQDADGTDPLGEIVYVSATLAADA--
Mvan_1344|M.vanbaalenii_PYR-1 TRYGRPLPPEAGDTDPLGEIVYVSAHRPQ----
MSMEG_1480|M.smegmatis_MC2_155 ADHGFELPEEMAELS--GDSGYLSAHLK-----
Mb3793c|M.bovis_AF2122/97 ARYGRSIPHSGEDSIPPNLFVSAQRATS-----
Rv3767c|M.tuberculosis_H37Rv ARYGRSIPHSGEDSIPPNLFVSAQRATS-----
MMAR_0955|M.marinum_M AGYGRAAAAQVEDRVPTNLFVAAQRK-------
MUL_0706|M.ulcerans_Agy99 AGYGRAAAAQVEDRVPTNLFVAAQRN-------
MAV_4558|M.avium_104 ARYDRRPPHDIEDATPQTRFVAAQRTERTRPDR
MAB_3886c|M.abscessus_ATCC_199 GRHGRDIPADLPDATPNSAFLFAQKAD------
. .