For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSTESGLDDVVAAAVRGDRDALSEVLSAIRPVVVRYCRARVGTAERSGLSADDVAQEVCLAVIAALPRYQ DQGRPFLAFVYGIAAHKVADAHRAMARDKSDPAENLPEQVDRHAGPEQEALNSESSRRMQALLEVLPEKQ REILILRVVVGLSAEETADAVGSSAGAVRVAQHRALQKLKAEMTRAGDYRG*
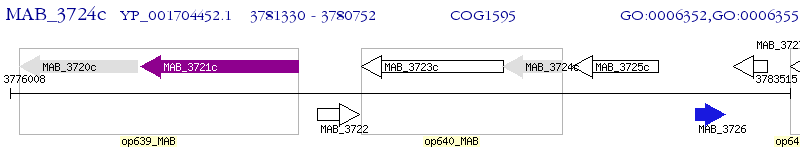
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3724c | - | - | 100% (192) | RNA polymerase sigma factor SigD |
| M. abscessus ATCC 19977 | MAB_3428c | - | 7e-12 | 32.39% (176) | RNA polymerase sigma-C factor |
| M. abscessus ATCC 19977 | MAB_1248 | - | 1e-10 | 27.75% (173) | RNA polymerase sigma factor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3448c | sigD | 6e-74 | 74.21% (190) | RNA polymerase sigma factor SigD |
| M. gilvum PYR-GCK | Mflv_4912 | - | 1e-75 | 75.39% (191) | RNA polymerase sigma factor SigD |
| M. tuberculosis H37Rv | Rv3414c | sigD | 6e-74 | 74.21% (190) | RNA polymerase sigma factor SigD |
| M. leprae Br4923 | MLBr_01448 | - | 2e-09 | 27.96% (186) | RNA polymerase sigma factor SigC |
| M. marinum M | MMAR_1134 | sigD | 8e-73 | 73.68% (190) | alternative RNA polymerase sigma-D factor, SigD |
| M. avium 104 | MAV_4359 | - | 3e-70 | 73.08% (182) | RNA polymerase sigma factor SigD |
| M. smegmatis MC2 155 | MSMEG_1599 | - | 4e-74 | 73.94% (188) | RNA polymerase sigma factor SigD |
| M. thermoresistible (build 8) | TH_1681 | sigD | 2e-71 | 74.58% (177) | PROBABLE ALTERNATIVE RNA POLYMERASE SIGMA-D FACTOR SIGD |
| M. ulcerans Agy99 | MUL_0897 | sigD | 5e-73 | 73.68% (190) | RNA polymerase sigma factor SigD |
| M. vanbaalenii PYR-1 | Mvan_1508 | - | 1e-75 | 76.60% (188) | RNA polymerase sigma factor SigD |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4912|M.gilvum_PYR-GCK ---------------------MTISGERLDAVVAEAVAGDRGALREVLET
Mvan_1508|M.vanbaalenii_PYR-1 ---------------------MTISGERLDAVVAEAVAGDRDALREVLET
MSMEG_1599|M.smegmatis_MC2_155 ---------------------MTSSGDRLDAVVAEAVAGDRDALREVLEI
TH_1681|M.thermoresistible__bu ------------------------------------VAGDRDALRHVLET
Mb3448c|M.bovis_AF2122/97 MVDPGVSPGCVRFVTLEISPSMTMQGERLDAVVAEAVAGDRNALREVLET
Rv3414c|M.tuberculosis_H37Rv MVDPGVSPGCVRFVTLEISPSMTMQGERLDAVVAEAVAGDRNALREVLET
MMAR_1134|M.marinum_M ---------------MEISPSMTIEGERLDAVVAEAVAGDRNALREVLET
MUL_0897|M.ulcerans_Agy99 ---------------MEISPSMTIEGERLDAVVAEAVAGDRNALREVLET
MAV_4359|M.avium_104 ---------------MTIP-----AGERLDAVVAKAVTGDHNALREVLET
MAB_3724c|M.abscessus_ATCC_199 ---------------------MTSTESGLDDVVAAAVRGDRDALSEVLSA
MLBr_01448|M.leprae_Br4923 -------------------MIASSGDEAVTDLALAAARGSKRALEAFIKA
. *.: ** .:.
Mflv_4912|M.gilvum_PYR-GCK IRPLVVRYCRARVGTAERSGLSADDVAQEVCLAAITALPRYRDQGRPFLA
Mvan_1508|M.vanbaalenii_PYR-1 IRPLVVRYCRARVGTAERSGLSADDVAQEVCLAAITALPRYKDQGRPFLA
MSMEG_1599|M.smegmatis_MC2_155 IRPIIVRYCRARIGATERSGLSADDVAQEVCLAAITALPRYRDQGRPFLA
TH_1681|M.thermoresistible__bu IRPLVLRYCRARVGAGERSGLSADDVAQEVCLAAITALPRYKDQGRPFLA
Mb3448c|M.bovis_AF2122/97 IRPIVVRYCRARVGTVERSGLSADDVAQEVCLATITALPRYRDRGRPFLA
Rv3414c|M.tuberculosis_H37Rv IRPIVVRYCRARVGTVERSGLSADDVAQEVCLATITALPRYRDRGRPFLA
MMAR_1134|M.marinum_M IRPIVVRYCRARVGTVERSGLSADDVAQEVCLATITALPRYRDRGRPFLA
MUL_0897|M.ulcerans_Agy99 IRPIVVRYCRARVGTVERSGLSADDVAQEVCLATITALPRYRDRGRPFLA
MAV_4359|M.avium_104 IRPIVVRYCRARVGTVERGGLSADDVAQEVCLATITALPRYRDRGRPFLA
MAB_3724c|M.abscessus_ATCC_199 IRPVVVRYCRARVGTAERSGLSADDVAQEVCLAVIAALPRYQDQGRPFLA
MLBr_01448|M.leprae_Br4923 TQQDVWRFVAYLSDVG-----NADDLTQETFLRAIGAIERFSGHS-SGRT
: : *: .. .***::**. * .* *: *: .:. . :
Mflv_4912|M.gilvum_PYR-GCK FVYGIAAHKVADAHRAAGRNRADPMDVVPERLSGEAGPEQMAIDSESSAR
Mvan_1508|M.vanbaalenii_PYR-1 FVYGIAAHKVADAHRAAGRNKADPMDVVPERFSGEAGPEQLAIDSESSAR
MSMEG_1599|M.smegmatis_MC2_155 FVYGIAAHKVADAHRAAARNRAEPTDVVPERFSLDAGPEQSAIDSESSAR
TH_1681|M.thermoresistible__bu FVYGIAAHKVADAHRAAARNRAEPTDVLPERISTENGPEQMAIDAESSAR
Mb3448c|M.bovis_AF2122/97 FLYGIAAHKVADAHRAAGRDRAYPAETLPERWSADAGPEQMAIEADSVTR
Rv3414c|M.tuberculosis_H37Rv FLYGIAAHKVADAHRAAGRDRAYPAETLPERWSADAGPEQMAIEADSVTR
MMAR_1134|M.marinum_M FLYGIAAHKVADAHRAAGRDLAYPAESVPERRSADAGPEQLAIEADSVTR
MUL_0897|M.ulcerans_Agy99 FLYGIAAHKVADAHRAAGRDLAYPAESVPERRSADAGPEQLAIEADSVTR
MAV_4359|M.avium_104 FLYGIAAHKVADAHRAAGRDLAYPTESIPDRWSNDAGPEQLAIEADSVSR
MAB_3724c|M.abscessus_ATCC_199 FVYGIAAHKVADAHRAMARDKSDPAENLPEQVDRHAGPEQEALNSESSRR
MLBr_01448|M.leprae_Br4923 WLLAIARRVVADHIRRAQTRPRTTFDGYPERMLN---ADRHARGFEDLVE
:: .** : *** * . : *:: .:: * :. .
Mflv_4912|M.gilvum_PYR-GCK MATLLKVLPEKQREILILRVVVGMSAEETAEAVGSTAGAVRVAQHRALSR
Mvan_1508|M.vanbaalenii_PYR-1 MAELLQILPEKQREILILRVVVGMSAEETADAVGSTAGAVRVAQHRALTR
MSMEG_1599|M.smegmatis_MC2_155 MAKLLSVLPEKQREILILRVVVGMSAEETAEAVGSTAGAVRVAQHRALAR
TH_1681|M.thermoresistible__bu MQKLLQVLPEKQREILILRVVVGMSAEETAEAVGSTAGAVRVAQHRALAR
Mb3448c|M.bovis_AF2122/97 MNELLEILPAKQREILILRVVVGLSAEETAAAVGSTTGAVRVAQHRALQR
Rv3414c|M.tuberculosis_H37Rv MNELLEILPAKQREILILRVVVGLSAEETAAAVGSTTGAVRVAQHRALQR
MMAR_1134|M.marinum_M MNELLEILPAKQREILILRVVVGLSAEETAAAVGSTTGAVRVAQHRALQR
MUL_0897|M.ulcerans_Agy99 MNELLEILPAKQREILILRVVVGLSAEETAAAVGSTTGAVRVAQHRALQR
MAV_4359|M.avium_104 MSELLEILPAKQREILILRVVVGLSAEETAAAVGSTTGAVRVAQHRALSR
MAB_3724c|M.abscessus_ATCC_199 MQALLEVLPEKQREILILRVVVGLSAEETADAVGSSAGAVRVAQHRALQK
MLBr_01448|M.leprae_Br4923 VTTMIANLATEQREALLLTQLLGLPYADAAAVCGCPVGTIRSRVARARDA
: :: *. :*** *:* ::*:. ::* . *...*::* **
Mflv_4912|M.gilvum_PYR-GCK LKAEITSTGQGRDYA-
Mvan_1508|M.vanbaalenii_PYR-1 LKTEIMAAG--RDHA-
MSMEG_1599|M.smegmatis_MC2_155 LKTEIMATG--RDHA-
TH_1681|M.thermoresistible__bu LKAEVMATG--HHHG-
Mb3448c|M.bovis_AF2122/97 LKDEIVAAG---DYA-
Rv3414c|M.tuberculosis_H37Rv LKDEIVAAG---DYA-
MMAR_1134|M.marinum_M LKDEIVAAG---DYA-
MUL_0897|M.ulcerans_Agy99 LKDEIVAAG---DYA-
MAV_4359|M.avium_104 LKSEMIAAG---DCA-
MAB_3724c|M.abscessus_ATCC_199 LKAEMTRAG---DYRG
MLBr_01448|M.leprae_Br4923 LLADTEHNH----LTG
* :