For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTILVTGATGNVGRPLVDLLVAAGADVRAVTQHPERASFPPQVATVASARAGLDGVTAVFLNSRALGEDL DRFVNQSAREGVTRLVALSAINCDDDFSRQPSRFRGDRNKEVEQYAAGSGLQWVSLRPTVFASNFLGMWG TQLQAGDVVRGPHAAASAAPIADSDISEVAARALLTDELLGQRIPLTGPQAFTNAGLVAVLGDVLGRPLE YQQVSDDLVRQRFGRLGFPAGFADAYLAMQAATVTEPAIVTHDVDRILGRPAETFASWANRFRTRFAPAE FAAQKG
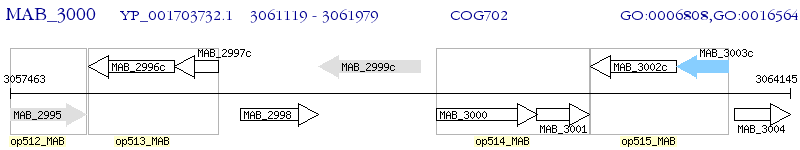
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3000 | - | - | 100% (286) | hypothetical protein MAB_3000 |
| M. abscessus ATCC 19977 | MAB_3360c | - | 2e-06 | 30.19% (212) | hypothetical protein MAB_3360c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2073c | - | 1e-07 | 26.76% (213) | hypothetical protein Mb2073c |
| M. gilvum PYR-GCK | Mflv_1635 | - | 8e-05 | 29.17% (144) | NAD-dependent epimerase/dehydratase |
| M. tuberculosis H37Rv | Rv2047c | - | 1e-07 | 26.76% (213) | hypothetical protein Rv2047c |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_4766 | - | 1e-06 | 29.00% (200) | hypothetical protein MMAR_4766 |
| M. avium 104 | MAV_0162 | - | 7e-98 | 66.30% (276) | hypothetical protein MAV_0162 |
| M. smegmatis MC2 155 | MSMEG_5366 | - | 1e-56 | 66.67% (159) | hypothetical protein MSMEG_5366 |
| M. thermoresistible (build 8) | TH_2433 | - | 1e-22 | 31.47% (286) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2101 | - | 1e-05 | 31.73% (208) | dTDP-4-dehydrorhamnose reductase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4766|M.marinum_M -----MKILVIGGSGLIGSQVVAQLTGLAHQAVAASPRS-----------
Mvan_2101|M.vanbaalenii_PYR-1 -----MKISVIGATGLIGSKVVDLLQRDGHHVVAASRAS-----------
Mb2073c|M.bovis_AF2122/97 -----MRIAVTGASGVLGRGLTARLLSQGHEVVGIARHRPDSWPSSA---
Rv2047c|M.tuberculosis_H37Rv -----MRIAVTGASGVLGRGLTARLLSQGHEVVGIARHRPDSWPSSA---
MAV_0162|M.avium_104 -----MTIVVTGATGNVGRPLVTALLAAGA-PVRAVTRRSGPAG------
MSMEG_5366|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3000|M.abscessus_ATCC_1997 -----MTILVTGATGNVGRPLVDLLVAAGA-DVRAVTQHPERAS------
TH_2433|M.thermoresistible__bu -----MTILVTGATGNIGRRVVDHLIALGATDIRALTTDPAKAK------
Mflv_1635|M.gilvum_PYR-GCK MPGNDVRCVVTGATGYIGGRLVPELLARGLQVRAMARTPSKLDRTEWRDR
MMAR_4766|M.marinum_M ----GVNALTGEGLADAVAGADAVVDVSNS--------------------
Mvan_2101|M.vanbaalenii_PYR-1 ----GADVLTGEGLTDALAGADVLVDVTNS--------------------
Mb2073c|M.bovis_AF2122/97 -DFIAADIRDATAVESAMTGADVVAHCAWVRGRNDHINIDGTANVLKAMA
Rv2047c|M.tuberculosis_H37Rv -DFIAADIRDATAVESAMTGADVVAHCAWVRGRNDHINIDGTANVLKAMA
MAV_0162|M.avium_104 ---FPAQVELFDTAADALPGASAVFLNAR---------------------
MSMEG_5366|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3000|M.abscessus_ATCC_1997 ---FPPQVATVASARAGLDGVTAVFLNSR---------------------
TH_2433|M.thermoresistible__bu ---LPESVTAVTGYLGRPETLPAALHGVRRM-------------------
Mflv_1635|M.gilvum_PYR-GCK VEVVRGDLTEADSLTAAFNGADVVYYLVHSMG------------------
MMAR_4766|M.marinum_M --------------------------------------------------
Mvan_2101|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 ETGTGRIVFTSSGHQPRVEQMLADCGLEWVAVRCALIFGRNVDNWVQRLF
Rv2047c|M.tuberculosis_H37Rv ETGTGRIVFTSSGHQPRVEQMLADCGLEWVAVRCALIFGRNVDNWVQRLF
MAV_0162|M.avium_104 --------------------------------------------------
MSMEG_5366|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3000|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2433|M.thermoresistible__bu --------------------------------------------------
Mflv_1635|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_4766|M.marinum_M --------------------------------------------------
Mvan_2101|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 ALPVLPAGYADRVVQVVHSDDAQRLLVRALLDTVIDSGPVNLAAPGELTF
Rv2047c|M.tuberculosis_H37Rv ALPVLPAGYADRVVQVVHSDDAQRLLVRALLDTVIDSGPVNLAAPGELTF
MAV_0162|M.avium_104 --------------------------------------------------
MSMEG_5366|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3000|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2433|M.thermoresistible__bu --------------------------------------------------
Mflv_1635|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_4766|M.marinum_M -------------------------------------------------P
Mvan_2101|M.vanbaalenii_PYR-1 -------------------------------------------------P
Mb2073c|M.bovis_AF2122/97 RRIAAALGRPMVPIGSPVLRRVTSFAELELLHSAPLMDVTLLRDRWGFQP
Rv2047c|M.tuberculosis_H37Rv RRIAAALGRPMVPIGSPVLRRVTSFAELELLHSAPLMDVTLLRDRWGFQP
MAV_0162|M.avium_104 -----------------------------------------------ALG
MSMEG_5366|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3000|M.abscessus_ATCC_1997 -----------------------------------------------ALG
TH_2433|M.thermoresistible__bu --------------------------------------------YLAPLP
Mflv_1635|M.gilvum_PYR-GCK ----------------------------------------TSRDFVAEER
MMAR_4766|M.marinum_M SFADDDVLHFFTTSTKNLLAAEQ------------------TAGVRHHVA
Mvan_2101|M.vanbaalenii_PYR-1 SFDDDAVMDFFTRSSANLVSAAE------------------ESGVRHYVA
Mb2073c|M.bovis_AF2122/97 AWNAEECLEDFTLAVRGRIGLGKRTFSLPWRLANIQDLPAVDSPADDGVA
Rv2047c|M.tuberculosis_H37Rv AWNAEECLEDFTLAVRGRIGLGKRTFSLPWRLANIQDLPAVDSPADDGVA
MAV_0162|M.avium_104 GQLEDVVARCARGGVTKLVALSAINADDDFSRQPSRFRGDRNREVEQLAV
MSMEG_5366|M.smegmatis_MC2_155 -------------------------------------------------M
MAB_3000|M.abscessus_ATCC_1997 EDLDRFVNQSAREGVTRLVALSAINCDDDFSRQPSRFRGDRNKEVEQYAA
TH_2433|M.thermoresistible__bu ATLPTTLELAEQAGVEYVVALSG---------------GAHWQQHADTIA
Mflv_1635|M.gilvum_PYR-GCK RSAHNVVEAAKKAGVRRIVYLSG-------------LHPDRKELSRHLAS
MMAR_4766|M.marinum_M LSIVG------------------------TDRVP----------------
Mvan_2101|M.vanbaalenii_PYR-1 LSIVG------------------------ADGLP----------------
Mb2073c|M.bovis_AF2122/97 PRLAGPEGANGEFDTPIDPRFPTYLATNLSEALPGPFSPSSASVTVRGLR
Rv2047c|M.tuberculosis_H37Rv PRLAGPEGANGEFDTPIDPRFPTYLATNLSEALPGPFSPSSASVTVRGLR
MAV_0162|M.avium_104 GSGLA-----------------------WVSLRPT---------------
MSMEG_5366|M.smegmatis_MC2_155 ESALQ-----------------------WVSLRPS---------------
MAB_3000|M.abscessus_ATCC_1997 GSGLQ-----------------------WVSLRPT---------------
TH_2433|M.thermoresistible__bu AAPLT-----------------------STQLGPG---------------
Mflv_1635|M.gilvum_PYR-GCK RVEVG-----------------------EILMQSG---------------
.
MMAR_4766|M.marinum_M --------------------------------------------------
Mvan_2101|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 AGGVGIAERLRPSGVIQREIAMRTVAVFAHRLYGAITSAHFMAATVPFAK
Rv2047c|M.tuberculosis_H37Rv AGGVGIAERLRPSGVIQREIAMRTVAVFAHRLYGAITSAHFMAATVPFAK
MAV_0162|M.avium_104 --------------------------------------------------
MSMEG_5366|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3000|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2433|M.thermoresistible__bu --------------------------------------------------
Mflv_1635|M.gilvum_PYR-GCK -------------------------------------------------I
MMAR_4766|M.marinum_M ------NSGYLRAKCAQEKLIKESDLPYSIVRATQFFEFV----------
Mvan_2101|M.vanbaalenii_PYR-1 ------ESGYMRAKVVQEKTIAGSDVPHSIVRATQFHEFA----------
Mb2073c|M.bovis_AF2122/97 PATIVSNSGFFGPSMASLPIFGAQRPPSESSRARRWLRTLRNIGVFGVNL
Rv2047c|M.tuberculosis_H37Rv PATIVSNSGFFGPSMASLPIFGAQRPPSESSRARRWLRTLRNIGVFGVNL
MAV_0162|M.avium_104 ----VFATNFAGMWSAQLREGDVVAGPYAAASSAPIVETD----------
MSMEG_5366|M.smegmatis_MC2_155 ----AFVSNFLGMWAPQLQAGDVVAGPYATASMAPIAEHD----------
MAB_3000|M.abscessus_ATCC_1997 ----VFASNFLGMWGTQLQAGDVVRGPHAAASAAPIADSD----------
TH_2433|M.thermoresistible__bu ----EFLENFT-LWAEQIKTTRTVREPYPDVVEAPISMDD----------
Mflv_1635|M.gilvum_PYR-GCK ETVVLQAGIVIGSGSASFEMVRHLTDRLPVMTTPKWVHNKIQP----ISI
.
MMAR_4766|M.marinum_M IGIADSATDGDTVRLSHTAAIQPIAASDVATGVTR---------------
Mvan_2101|M.vanbaalenii_PYR-1 DAIVGTLEADDEVRVP-DARIQPIAADDVAAEVFR---------------
Mb2073c|M.bovis_AF2122/97 VGLSAGSPRDTDAYVADVDRLERLAFDNLATHDDRRLLSLILLARDHVVH
Rv2047c|M.tuberculosis_H37Rv VGLSAGSPRDTDAYVADVDRLERLAFDNLATHDDRRLLSLILLARDHVVH
MAV_0162|M.avium_104 IADVAAHALLTDD--LVGQRIPLTGPQALSN-------------------
MSMEG_5366|M.smegmatis_MC2_155 IAAVAAHALLTDD--LVGQKVPLTGPQALTN-------------------
MAB_3000|M.abscessus_ATCC_1997 ISEVAARALLTDE--LLGQRIPLTGPQAFTN-------------------
TH_2433|M.thermoresistible__bu IARVAAHLLAEPDPSHHGRMYELTGPQALTR-------------------
Mflv_1635|M.gilvum_PYR-GCK SDVLHYLAEAATAEFGESRTWDIGGPDVMEYG------------------
. . .
MMAR_4766|M.marinum_M ------------AAISDPLNGTTN-----IAGPDKFGMDDFVRMG-----
Mvan_2101|M.vanbaalenii_PYR-1 ------------AAVGSPVDGVIN-----IGGPDKISFADLARAV-----
Mb2073c|M.bovis_AF2122/97 GWVLASGSFMLCAAFNVLLRGLCGRDTAPAAGPELVSARSVEAVQRLVAA
Rv2047c|M.tuberculosis_H37Rv GWVLASGSFMLCAAFNVLLRGLCGRDTAPAAGPELVSARSVEAVQRLVAA
MAV_0162|M.avium_104 ------------AQLVEVIGTVLGRSLRYREIPAAAVRRRFVGLGLG---
MSMEG_5366|M.smegmatis_MC2_155 ------------ADLVDIIGGVLGRPLRYQEIPAVAVRQRFIALGFP---
MAB_3000|M.abscessus_ATCC_1997 ------------AGLVAVLGDVLGRPLEYQQVSDDLVRQRFGRLGFP---
TH_2433|M.thermoresistible__bu ------------ARIAEQIGVGIGVDVTFRRCDRAEAERALEPVMG----
Mflv_1635|M.gilvum_PYR-GCK ----------DAMQGYADVAGLRRRVMVVLPFLTPTIASWWVGLVTP---
:
MMAR_4766|M.marinum_M ----------LAAHGDSRQVRTDPNARYFGAVLDGHSIVPTDGE-DVTIY
Mvan_2101|M.vanbaalenii_PYR-1 ----------LARRGADKTVVIDPQATYFGTPLDQNSLVTGDG---AVLG
Mb2073c|M.bovis_AF2122/97 ARRDPVVIRLLAEPGERLDKLAVEAPEFHSAVLAELTLIGHRGPAEVEMA
Rv2047c|M.tuberculosis_H37Rv ARRDPVVIRLLAEPGERLDKLAVEAPEFHSAVLAELTLIGHRGPAEVEMA
MAV_0162|M.avium_104 -----------AGFADAYLAMLAETLDKPALVTHDVEKILGRPAVPFAHW
MSMEG_5366|M.smegmatis_MC2_155 -----------AAFADAYIQLLAATVDKPALVTHDVEKILGRPATTFAEA
MAB_3000|M.abscessus_ATCC_1997 -----------AGFADAYLAMQAATVTEPAIVTHDVDRILGRPAETFASW
TH_2433|M.thermoresistible__bu ------------ARARWYLDLLAD--GAPQRANRLVEELTGTPAESVAQW
Mflv_1635|M.gilvum_PYR-GCK ---------IPSGLARPLVESLECDAICHEHDIDAVIPPPKGGLIGYREA
.
MMAR_4766|M.marinum_M PTHFSDWLAAG---------------------------------------
Mvan_2101|M.vanbaalenii_PYR-1 AVRFADHH------------------------------------------
Mb2073c|M.bovis_AF2122/97 ATSYADNPELLVRMVAKTLRAVPAPQPPTPVIPLRAKPVALLAARQLRDR
Rv2047c|M.tuberculosis_H37Rv ATSYADNPELLVRMVAKTLRAVPAPQPPTPVIPLRAKPVALLAARQLRDR
MAV_0162|M.avium_104 VATHRDLFDRRNERKEGV--------------------------------
MSMEG_5366|M.smegmatis_MC2_155 VSTHRDLFAKH---------------------------------------
MAB_3000|M.abscessus_ATCC_1997 ANRFRTRFAPAEFAAQKG--------------------------------
TH_2433|M.thermoresistible__bu ATRHAELFR-----------------------------------------
Mflv_1635|M.gilvum_PYR-GCK VAKELRSEGGSSAGRRHLLRFSPAASD-----------------------
MMAR_4766|M.marinum_M --------------------------------------------------
Mvan_2101|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 EVRRDRMVRAIWVLRALLREYGRRLTEAGVFDTPDDVFYLLVDEIDALPA
Rv2047c|M.tuberculosis_H37Rv EVRRDRMVRAIWVLRALLREYGRRLTEAGVFDTPDDVFYLLVDEIDALPA
MAV_0162|M.avium_104 --------------------------------------------------
MSMEG_5366|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3000|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2433|M.thermoresistible__bu --------------------------------------------------
Mflv_1635|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_4766|M.marinum_M --------------------------------------------------
Mvan_2101|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 DVSGLVARRRAEQRRLAGIVPPTVFSGSWEPSPSSAAALAAGDTLRGVGV
Rv2047c|M.tuberculosis_H37Rv DVSGLVARRRAEQRRLAGIVPPTVFSGSWEPSPSSAAALAAGDTLRGVGV
MAV_0162|M.avium_104 --------------------------------------------------
MSMEG_5366|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3000|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2433|M.thermoresistible__bu --------------------------------------------------
Mflv_1635|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_4766|M.marinum_M --------------------------------------------------
Mvan_2101|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 CGGRVRGRVRIVRPETIDDLQPGEILVAEVTDVGYTAAFCYAAAVVTELG
Rv2047c|M.tuberculosis_H37Rv CGGRVRGRVRIVRPETIDDLQPGEILVAEVTDVGYTAAFCYAAAVVTELG
MAV_0162|M.avium_104 --------------------------------------------------
MSMEG_5366|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3000|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2433|M.thermoresistible__bu --------------------------------------------------
Mflv_1635|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_4766|M.marinum_M --------------------------------------------------
Mvan_2101|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2073c|M.bovis_AF2122/97 GPMSHAAVVAREFGFPCVVDAQGATRFLPPGALVEVDGATGEIHVVELAS
Rv2047c|M.tuberculosis_H37Rv GPMSHAAVVAREFGFPCVVDAQGATRFLPPGALVEVDGATGEIHVVELAS
MAV_0162|M.avium_104 --------------------------------------------------
MSMEG_5366|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3000|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2433|M.thermoresistible__bu --------------------------------------------------
Mflv_1635|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_4766|M.marinum_M -------------
Mvan_2101|M.vanbaalenii_PYR-1 -------------
Mb2073c|M.bovis_AF2122/97 EDGPALPGSDLSR
Rv2047c|M.tuberculosis_H37Rv EDGPALPGSDLSR
MAV_0162|M.avium_104 -------------
MSMEG_5366|M.smegmatis_MC2_155 -------------
MAB_3000|M.abscessus_ATCC_1997 -------------
TH_2433|M.thermoresistible__bu -------------
Mflv_1635|M.gilvum_PYR-GCK -------------