For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KITVVGATGQIGSRVVSLLIADGHDVVAASLSSGANVLTGEGLVDALTGSAVVIDVVNSPSFEDGPVMDF FTASARNLVDAAKATGVGHYVALSIVGADGLPESGYMRAKVAQEKIIVGSGLPYTIVRATQFQEFAEAIT DTLVVGDEVRVPDARIQLIAVDDVSTQVAHAAAAAPRNGIINIGGPEKFSFADMAGAVLAARGDDRPVVV DSGATYFGTPVDDFSLVTGDDGVLTQTRFADWMARR*
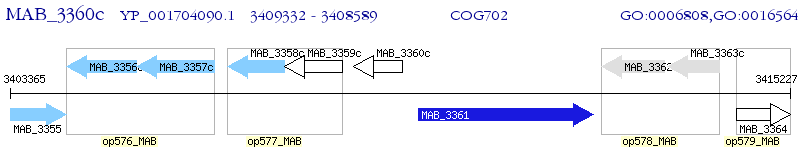
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3360c | - | - | 100% (247) | hypothetical protein MAB_3360c |
| M. abscessus ATCC 19977 | MAB_3000 | - | 2e-06 | 30.19% (212) | hypothetical protein MAB_3000 |
| M. abscessus ATCC 19977 | MAB_1344c | - | 1e-05 | 26.06% (165) | putative dTDP-glucose-4,6-dehydratase-related protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1635 | - | 4e-11 | 27.85% (237) | NAD-dependent epimerase/dehydratase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_4766 | - | 6e-63 | 52.82% (248) | hypothetical protein MMAR_4766 |
| M. avium 104 | MAV_4078 | - | 2e-20 | 35.51% (214) | hypothetical protein MAV_4078 |
| M. smegmatis MC2 155 | MSMEG_2353 | - | 3e-96 | 72.47% (247) | secreted protein |
| M. thermoresistible (build 8) | TH_3570 | - | 2e-84 | 64.90% (245) | PUTATIVE secreted protein |
| M. ulcerans Agy99 | MUL_0982 | - | 4e-08 | 28.09% (235) | hypothetical protein MUL_0982 |
| M. vanbaalenii PYR-1 | Mvan_2101 | - | 6e-89 | 69.01% (242) | dTDP-4-dehydrorhamnose reductase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3360c|M.abscessus_ATCC_199 -------------------------------------MKITVVGATGQIG
MSMEG_2353|M.smegmatis_MC2_155 -------------------------------------MKITVFGATGLIG
Mvan_2101|M.vanbaalenii_PYR-1 -------------------------------------MKISVIGATGLIG
TH_3570|M.thermoresistible__bu --------------------------------VEGAVMRITVFGATGRIG
MMAR_4766|M.marinum_M -------------------------------------MKILVIGGSGLIG
Mflv_1635|M.gilvum_PYR-GCK ------------------MPG--------------NDVRCVVTGATGYIG
MUL_0982|M.ulcerans_Agy99 MCSCGQSRCRFRIGSDRLLPDGPASPRPHRLGSSVETMRILVTGATGYVG
MAV_4078|M.avium_104 ---------------------------------MRALTTIAVIGATGTAG
* *.:* *
MAB_3360c|M.abscessus_ATCC_199 SRVVSLLIADGHDVVAASLSSG--------------ANVLTGEGLVDALT
MSMEG_2353|M.smegmatis_MC2_155 SKVVAALNEAGHDVVAAARDTG--------------ADVLTGEGLADALT
Mvan_2101|M.vanbaalenii_PYR-1 SKVVDLLQRDGHHVVAASRASG--------------ADVLTGEGLTDALA
TH_3570|M.thermoresistible__bu SEVVALLRADGHDVVPASLSTG--------------ADVVTGAGLADSLS
MMAR_4766|M.marinum_M SQVVAQLTGLAHQAVAASPRSG--------------VNALTGEGLADAVA
Mflv_1635|M.gilvum_PYR-GCK GRLVPELLARGLQVRAMARTPSKLDRTEWRDRVEVVRGDLTEADSLTAAF
MUL_0982|M.ulcerans_Agy99 SRLVTALLADGHEVLAATRNMARLSRLAWFDDVTPVILDATDRASAQAAM
MAV_4078|M.avium_104 SRVVARLRPRDVAVVEISRAQG--------------VDVFDAPGLTRALE
..:* * . : . :
MAB_3360c|M.abscessus_ATCC_199 GSAVVIDVVNSPSFEDGPVMDFFT---ASARNLVDAAKATGVGHYVALSI
MSMEG_2353|M.smegmatis_MC2_155 GAEVLVDVVNSPSFADDDVMNFFT---TSTRNLTEAATKTGTGHYVALSI
Mvan_2101|M.vanbaalenii_PYR-1 GADVLVDVTNSPSFDDDAVMDFFT---RSSANLVSAAEESGVRHYVALSI
TH_3570|M.thermoresistible__bu GADVLVDVVNSPSFADGPVSEFFT---AAALNLTAAARAAGVGHHVALSI
MMAR_4766|M.marinum_M GADAVVDVSNSPSFADDDVLHFFT---TSTKNLLAAEQTAGVRHHVALSI
Mflv_1635|M.gilvum_PYR-GCK NGA---DVVYYLVHSMGTSRDFVAEERRSAHNVVEAAKKAGVRRIVYLSG
MUL_0982|M.ulcerans_Agy99 NAAGQIDVVYYLVHGIGQP-DFRDRDKTAAANLAVAARDTGVRRIVYLGG
MAV_4078|M.avium_104 GVDVVIDVSN--PVPPDGRSDIRQTLATAARNIMGACATQEVQRLVVLTI
. ** . .: :: *: * . : * *
MAB_3360c|M.abscessus_ATCC_199 VGADG--LPESGYMRAKVAQEKIIVGS--------GLPYTIVRATQFQEF
MSMEG_2353|M.smegmatis_MC2_155 VGCDA--LPDSGYMRAKVAQEKAIAAS--------GIDHTIVRATQFHEF
Mvan_2101|M.vanbaalenii_PYR-1 VGADG--LPESGYMRAKVVQEKTIAGS--------DVPHSIVRATQFHEF
TH_3570|M.thermoresistible__bu VGCDE--LPDSGYMRAKAAQERIIAES--------GLPYTIVRATQFDEF
MMAR_4766|M.marinum_M VGTDR--VPNSGYLRAKCAQEKLIKES--------DLPYSIVRATQFFEF
Mflv_1635|M.gilvum_PYR-GCK LHPDR--KELSRHLASRVEVGEILMQS-GIETVVLQAGIVIGSGSASFEM
MUL_0982|M.ulcerans_Agy99 FVPEC--EALSQHLSSRAVVAEALAVDDGAELVWLGAAMIIGAGSTLFEM
MAV_4078|M.avium_104 AGIEDPVFDEFPYYQAKREAKEILLDG--------PVPTTVVKSTQWYEF
: : :: . : . : .: *:
MAB_3360c|M.abscessus_ATCC_199 AEAITDTLVVGDEVRVPD-ARIQLIAVDDVSTQVAHAAAAAPR-NGIINI
MSMEG_2353|M.smegmatis_MC2_155 AESITQTLVAGDEVRVPD-GRIQLIASDEVAAEVARAAQAAAV-NGIVNI
Mvan_2101|M.vanbaalenii_PYR-1 ADAIVGTLEADDEVRVPD-ARIQPIAADDVAAEVFRAAVGSPV-DGVINI
TH_3570|M.thermoresistible__bu AEAIVESLIVGAEVRVPD-GSIQPIAAADVAAEVAARAVAEPA-GGVVNI
MMAR_4766|M.marinum_M VIGIADSATDGDTVRLSHTAAIQPIAASDVATGVTRAAISDPL-NGTTNI
Mflv_1635|M.gilvum_PYR-GCK VRHLTDRLPVMTTPKWVH-NKIQPISISDVLHYLAEAATAEFGESRTWDI
MUL_0982|M.ulcerans_Agy99 LRYVGDRFPVLPMPDWMY-NPIDPISIRDVLHYLVAAADVDQVPAGAYDI
MAV_4078|M.avium_104 ATNHDAVSCDDNEVIVED-WLIQPIAADTVADVLVETALAQTH--TPRTI
*: *: * : * *
MAB_3360c|M.abscessus_ATCC_199 GGPEKFSFADMAGAVLAARGDDRPVVVDS-----------------GATY
MSMEG_2353|M.smegmatis_MC2_155 GGPEKLSFADMARAVLAKHGDDKPVVVDA-----------------QATY
Mvan_2101|M.vanbaalenii_PYR-1 GGPDKISFADLARAVLARRGADKTVVIDP-----------------QATY
TH_3570|M.thermoresistible__bu GGPDRMSFADLARTVLAHQHRRTPVVIDP-----------------QATY
MMAR_4766|M.marinum_M AGPDKFGMDDFVRMGLAAHGDSRQVRTDP-----------------NARY
Mflv_1635|M.gilvum_PYR-GCK GGPDVMEYGDAMQGYADVAGLRRRVMVVLPFLTPTIASWWVG--LVTPIP
MUL_0982|M.ulcerans_Agy99 CGPDTTSYRELLKTYARIRG-RWHAALPVGRLDTAVASRITAGHPAGPAW
MAV_4078|M.avium_104 TGPDAIRLPELTSKLLVRQGDRRAVRSVEP----------------AVSA
**: : .
MAB_3360c|M.abscessus_ATCC_199 FG------TPVDDFSLVT--GDDGVLTQTRFADWMARR------------
MSMEG_2353|M.smegmatis_MC2_155 FG------TAVDDDSLVT--GDDGKLATIRFADWLAGR------------
Mvan_2101|M.vanbaalenii_PYR-1 FG------TPLDQNSLVT--GDGAVLGAVRFADHH---------------
TH_3570|M.thermoresistible__bu FG------TRIDDTSLVT--GAGAVIAGTPLAAVLAPPASD---------
MMAR_4766|M.marinum_M FG------AVLDGHSIVPTDGEDVTIYPTHFSDWLAAG------------
Mflv_1635|M.gilvum_PYR-GCK SGLARPLVESLECDAICHEHDIDAVIPPPKGGLIGYREAVAKELRSEGGS
MUL_0982|M.ulcerans_Agy99 SGRRPGGIAGSPDGGIRGRPARQGPRATRRTTEYRRCHYVGAGRPRERAP
MAV_4078|M.avium_104 LG---AGALLASGHAVVVGPDVDTWLQTLAPPGGDGHRGRDGDGHRGADG
* .:
MAB_3360c|M.abscessus_ATCC_199 ----------------------
MSMEG_2353|M.smegmatis_MC2_155 ----------------------
Mvan_2101|M.vanbaalenii_PYR-1 ----------------------
TH_3570|M.thermoresistible__bu ----------------------
MMAR_4766|M.marinum_M ----------------------
Mflv_1635|M.gilvum_PYR-GCK SAGRR----HLLRFSPAASD--
MUL_0982|M.ulcerans_Agy99 VARRRPDRSPPTRIHRSGVGRR
MAV_4078|M.avium_104 DHRGADGDGKPRRG--------