For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AIRSVSALVLDNLAMFEFGVICEVFGIDRSADGVPNFDFKVCGPEAGSPLRTSVGATIIPDHGLDHLVGA DLVAIPAIGGSDYLPEALDAVRRAAESGSIILTVCSGAFVAGAAGLLDGRPCTTHWMHADELARMYPTAK VDRNVLFVDDGNLITSAGTAAGIDACLHLVRRELGSEVTNKIARRMVVPPQRDGGQRQYIDQPIPVKCSE RFAPHLDWILANLDKPHTVTTLARRAHMSGRTFARRFVEETGRTPMQWVTDQRVLFARRMLEETNLDIDR IAEQSGFGTATLLRHHFRRIIGVTPSDYRRRFNSTASQDVAVIA*
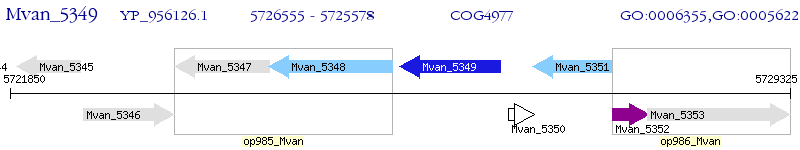
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5349 | - | - | 100% (325) | AraC family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5829 | - | 9e-44 | 40.57% (244) | AraC family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0972 | - | 5e-39 | 38.25% (285) | AraC family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1966c | - | 2e-16 | 30.57% (265) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_1437 | - | 1e-169 | 89.60% (327) | AraC family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1931c | - | 2e-16 | 30.57% (265) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0557 | - | 1e-129 | 71.15% (312) | AraC family transcriptional regulator |
| M. marinum M | MMAR_2855 | - | 4e-42 | 38.24% (272) | transcriptional regulatory protein |
| M. avium 104 | MAV_4981 | - | 4e-38 | 33.44% (323) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_5193 | - | 1e-74 | 44.86% (321) | transcriptional regulator, AraC family protein |
| M. thermoresistible (build 8) | TH_3120 | - | 1e-146 | 81.82% (308) | PUTATIVE transcriptional regulator, AraC family with |
| M. ulcerans Agy99 | MUL_2902 | - | 1e-40 | 37.50% (272) | transcriptional regulatory protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5349|M.vanbaalenii_PYR-1 -----MAIRSVSALVLDNLAMFEFGVICEVFGIDRSADGVPNFDFKVCGP
Mflv_1437|M.gilvum_PYR-GCK -----MAIKAVSTLVLDGLAVFEFGVVCEVFGIDRSADGVPNFDFKVCGP
TH_3120|M.thermoresistible__bu ----------VAALVLDGLAIFEFGVICEVFGVDRTDDGVPPVDFTVCGP
MAB_0557|M.abscessus_ATCC_1997 ------MLRSVAALVLDRVAVFEFGVICEVFGIDRARDGVPNFDFRVCGV
MSMEG_5193|M.smegmatis_MC2_155 -------MHRVAVLLIPPVIAFDATIAPMLFGYATDADGNPLYEVVTCGV
Mb1966c|M.bovis_AF2122/97 ----------MVIVGFPG--------------------------------
Rv1931c|M.tuberculosis_H37Rv ----------MVIVGFPG--------------------------------
MMAR_2855|M.marinum_M ------MARKVVIVGFPGVQPLDVVGPFEVFAGASLLSE--GGYDVALAS
MUL_2902|M.ulcerans_Agy99 ------MARKVVTVGFPGVQPLDVVGPFEVFAGASLLSE--GGYDVALAS
MAV_4981|M.avium_104 MADSSATPRVVVIVVFDDVTMLDVAGAGEVFAEANRFG---ADYLLKIAS
: : :
Mvan_5349|M.vanbaalenii_PYR-1 EAGSPLRTSVGATIIPDHGLDHLVGADLVAIP----AIGGSD--YLPEAL
Mflv_1437|M.gilvum_PYR-GCK EPGEPLRTSVGATITPDHGLDDLVGADLVAIP----AIASRQGTYLPEAL
TH_3120|M.thermoresistible__bu RAGQPVRTSFGATLTPDSGLDGLAGADLVAIP----AIRQDD--YLPEAL
MAB_0557|M.abscessus_ATCC_1997 EPGVPLRTTVGASLIPDHGLEGLADADLVALP----ARPFTEG-YPQEAL
MSMEG_5193|M.smegmatis_MC2_155 D-GEPVHSTNGFAIVPAAGPEALASADTVVVPGTTYAAARQDG-VLEEDV
Mb1966c|M.bovis_AF2122/97 -----------------------DPVDTVILPG---GAGVDAARSEPALI
Rv1931c|M.tuberculosis_H37Rv -----------------------DPVDTVILPG---GAGVDAARSEPALI
MMAR_2855|M.marinum_M IDERPVATGIGLEFMTTRLPDPSAPVDTVVLPG---GGGVDAARDNVALM
MUL_2902|M.ulcerans_Agy99 IDERPVATGIGLEFMTTRLPDPSAPVDTVVLPG---GGGVDAARDNVALM
MAV_4981|M.avium_104 VDGRDVTTSIGTRLGVTDAVSSIESADTVLVAG---SDNLPRRPIDPELV
.* * :. . :
Mvan_5349|M.vanbaalenii_PYR-1 DAVRRAAESGSIILTVCSGAFVAGAAGLLDGRPCTTHWMHADELARMYPT
Mflv_1437|M.gilvum_PYR-GCK DAIRRAAESGSIILTVCSGAFVAGAAGLLDGRPCTTHWMHADALAEMYPT
TH_3120|M.thermoresistible__bu DAVRAAADAGAIILTVCSGAFLAGAAGLLDGRPCTTHWGLTEELARRYPT
MAB_0557|M.abscessus_ATCC_1997 DAVRAASERGATILTVCSGAFIAGAAGLLDGRRCTTHWMYTDTLARQYPT
MSMEG_5193|M.smegmatis_MC2_155 SAALARIPAGTRLVSICTGAFVLAAAGLLDGRPATTHWRWADELRRLHPK
Mb1966c|M.bovis_AF2122/97 DWVKAVSGTARRVVTVCTGAFLAAEAGLLG--RTPSDDALG--LCRTFRP
Rv1931c|M.tuberculosis_H37Rv DWVKAVSGTARRVVTVCTGAFLAAEAGLLG--RTPSDDALG--LCRTFRP
MMAR_2855|M.marinum_M DWVKAAAGTARRVVTVCTGTFLAAEAGLLDGCRATTHWAFADRLAREFPA
MUL_2902|M.ulcerans_Agy99 DWVKAAAGTARRVVTVCTGTFLAAEAGLLDGCRATTHWAFADRLAREFPA
MAV_4981|M.avium_104 EAVKSVADRTRRLASICTGSFILGQAGLLDGRRATTHWHDVRLFARAFPE
. : ::*:*:*: . ****. .:. : . .
Mvan_5349|M.vanbaalenii_PYR-1 AKVDRNVLFVDDG-NLITSAGTAAGIDACLHLVRRELGSEVTNKIARRMV
Mflv_1437|M.gilvum_PYR-GCK AKVDRNVLFVDDG-NLITSAGTAAGIDACLHLVRREMGSEVTNKIARRMV
TH_3120|M.thermoresistible__bu ARVDRNVLYVDDG-DLITSAGTAAGIDACLHLVRRELGSEVTNTIARHMV
MAB_0557|M.abscessus_ATCC_1997 ATVDRDVLFVDDG-DLITSAGTAAGIDACLHLVRRELGSEITNRIARRMV
MSMEG_5193|M.smegmatis_MC2_155 IAVDENVLFVDDG-DLLTSAGLAAGIDLCLHIIRTDHGAGVANAVARYCV
Mb1966c|M.bovis_AF2122/97 R-ISGRSGRCRPDLHAQFAEGVDRGWSHRRHRPRAGTGRRRPRHRDCPDG
Rv1931c|M.tuberculosis_H37Rv R-ISGRSGRCRPDLHAQFAEGVDRGWSHRRHRPRAGTGRRRPRHRDCPDG
MMAR_2855|M.marinum_M VDVDPDPIFIRSSPKVWTAAGVTAGIDLALALVEGDHGTEIAQTVARWLV
MUL_2902|M.ulcerans_Agy99 VDVDPDPIFIRSSPKVWTAAGVTAGIDLALALVEGDHGTEIAQTVARWLV
MAV_4981|M.avium_104 ITVEPDALFVCDG-DVFTSAGISSGIDLALALVEMDYGTELVRAVARWLV
:. . . : * * . . * .
Mvan_5349|M.vanbaalenii_PYR-1 VPPQRDGGQ----RQYIDQ--PIPVKCSERFAPHLDWILANLDKPHTVTT
Mflv_1437|M.gilvum_PYR-GCK VPPQRDGGQ----RQYIDQ--PIPVKCSERFAPQLDWILANLDKPHTVTS
TH_3120|M.thermoresistible__bu VPPQRDGGQ----RQFIDQ--PIPVRHSEGFAPHLDWILSNLDKTHTVAS
MAB_0557|M.abscessus_ATCC_1997 VPPQRAGGQ----RQFIPQ--PVPDMVADGFGELCDWLIENIDQSHTVAE
MSMEG_5193|M.smegmatis_MC2_155 VPPWREGGQ----AQFIDRRLPEPEQATTSTAGTREWALDRLHEELTVER
Mb1966c|M.bovis_AF2122/97 CPLARPVSAPTRWADPVRGSGVDATRQTDLDPPGAGGHRGRAGGAHRIGE
Rv1931c|M.tuberculosis_H37Rv CPLARPVSAPTRWADPVRGSGVDATRQTDLDPPGAGGHRGRAGGAHRIGE
MMAR_2855|M.marinum_M LYLRRPGGQ-TQFAAPVWMP--RAKRPSIRDVQEA--IEAQPGGPHSIEE
MUL_2902|M.ulcerans_Agy99 LYLRRPCGQ-TQFAAPVWMP--RAKRPSIRDVQEA--IEAQPGGPHSIEE
MAV_4981|M.avium_104 VYLKRAGGQ-SQFSALVEAE--PPPRSPLRKVTAA--ISADPAADHSVKS
* . : . . :
Mvan_5349|M.vanbaalenii_PYR-1 LARRAHMSGRTFARRFVEETGRTPMQWVTDQRVLFARRMLEETNLDIDRI
Mflv_1437|M.gilvum_PYR-GCK LSRRAHMSGRTFARRFVEETGRTPMQWITDQRVLYARRMLEETTLDIDRI
TH_3120|M.thermoresistible__bu LARRANMSPRTFARRFVEETGRTPMQWVTDQRVLYARRLLEETDLDIHRI
MAB_0557|M.abscessus_ATCC_1997 LAARVHMSTRTFARKFTDQTGVTPMRWIIDQRVLRARRLLEETGLDIDTV
MSMEG_5193|M.smegmatis_MC2_155 LARHAHMSARTFSRRFAEETGQSPGTWIRNRRIDYARELLESHDLPIDEV
Mb1966c|M.bovis_AF2122/97 LAQRAAMSPRHFTRVFSDEVGEAPGRYVERIRTEAARRQLEETHDTVVAI
Rv1931c|M.tuberculosis_H37Rv LAQRAAMSPRHFTRVFSDEVGEAPGRYVERIRTEAARRQLEETHDTVVAI
MMAR_2855|M.marinum_M LAHRAAMSPRHFTRVFTGEVGEAPRQYVERVRTEAARRQLEETEDTVVAI
MUL_2902|M.ulcerans_Agy99 LAHRAAMSPRHFTRVFTGEVGEAPGRYVERVRTEAARRQLEETEDTVVAI
MAV_4981|M.avium_104 LSARASLSTRQLTRLFQSELGTTPARYVEMVRIDAARAALDAGR-SVAES
*: :. :* * ::* * : * :* :: * ** *: :
Mvan_5349|M.vanbaalenii_PYR-1 AEQSGFGTATLLRHHFRRIIGVTPSDYRRRFN-----STASQDVAVIA-
Mflv_1437|M.gilvum_PYR-GCK AEQSGFGTATLLRHHFRRLIGVTPSDYRKRFN-----AHAP-EVAEIA-
TH_3120|M.thermoresistible__bu AARCGFGSTTLLRHHFRRVIGVTPSDYRRRFACGSTTSGSTESGAEVTD
MAB_0557|M.abscessus_ATCC_1997 AARSGFGTAILLRHHFRRGVGITPTDYRKAFA-----------VAAR--
MSMEG_5193|M.smegmatis_MC2_155 ARLSGLGTAGNLRHHLRRGLGMSPSSYRKVYQ------------GT---
Mb1966c|M.bovis_AF2122/97 AARCGFGTAETMRRSFIRRVGISPDQYRKAFA-----------------
Rv1931c|M.tuberculosis_H37Rv AARCGFGTAETMRRSFIRRVGISPDQYRKAFA-----------------
MMAR_2855|M.marinum_M AARCGFGTAETMRRNFIRRVGISPDQYRKAFA-----------------
MUL_2902|M.ulcerans_Agy99 AARCGFGTAETMRRNFIRRVGISPDQYRKAFA-----------------
MAV_4981|M.avium_104 ARLAGFGSAETLRRVFVEHLGLSPKAYRDRFRT-----------AARG-
* .*:*:: :*: : . :*::* ** :