For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDPQSRPALMTRRTVLSAGAMLALGSTSLIGCSPVLPNEIPAGVRWEDLRRSLSGALVVRGDAAMDESGR AFNPLFDVNHPGAVAFCASDQDVARCVEFATSARLPIAARGGGHSYAAYCIPNDGLVVDLARMAAVSVTG TRAVVGAGARLIDVYAGIAGAGRMLAGGSCPTVGIAGLTLGGGVGVLTRRFGLTCDQLISARVVTADGKI RVVSADTESDLFWAIRGVGGGNFCIATELAFETAASTDLTVFTLDYAAGEMAPIVHRWLTFMAGAPDELW TTLHAIGGAIPQCRIVGCVAQGVNSQDVIESLRGGIGVRAADSFIAEMTFLDAMKFMGGCTTLTVAQCHP SWTGPGSGQLKREAFVASSRMVPHPDVDTARIETLLAGKPGLTFIFDSLGGAVRRISPDATAFPHRQAAA CIQIYHGVGADPAVAHERVSQARDGLGDICGPAAYVNYIDPGMPDWATAYYGDNLPRLRGIAAAYDPKGI FRFAQAVRP
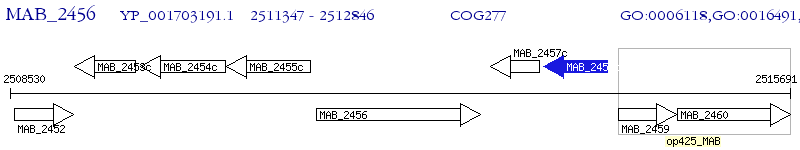
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2456 | - | - | 100% (499) | hypothetical protein MAB_2456 |
| M. abscessus ATCC 19977 | MAB_1407c | - | 7e-07 | 28.40% (162) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0064 | - | 7e-53 | 30.18% (487) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4651 | - | 3e-26 | 27.63% (485) | FAD linked oxidase domain-containing protein |
| M. tuberculosis H37Rv | Rv0063 | - | 7e-53 | 30.18% (487) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1179 | - | 1e-58 | 31.58% (494) | oxidoreductase |
| M. avium 104 | MAV_0089 | - | 3e-51 | 29.05% (482) | FAD binding domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1746 | - | 2e-29 | 28.64% (433) | 6-hydroxy-D-nicotine oxidase |
| M. thermoresistible (build 8) | TH_1540 | - | 1e-18 | 29.67% (182) | PROBABLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_1431 | - | 3e-58 | 31.38% (494) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_4048 | - | 3e-45 | 28.89% (495) | FAD linked oxidase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0064|M.bovis_AF2122/97 -----MAREISRQTFLRGAAGALAAG---AVFGSVRATADPAASGWEALS
Rv0063|M.tuberculosis_H37Rv -----MAREISRQTFLRGAAGALAAG---AVFGSVRATADPAASGWEALS
MAV_0089|M.avium_104 -----MAREISRQTFLRGAVGALAAG---AMLGAPRVGADPRPSGWEGLS
MMAR_1179|M.marinum_M --------MISRQTLLRGAAAATAAGLGSALFGSARAVAEP-VSGWSALA
MUL_1431|M.ulcerans_Agy99 --------MISRQTFLRGAAAATAAGLGSALFGSARAVAEP-VSGWSALA
MAB_2456|M.abscessus_ATCC_1997 MDPQSRPALMTRRTVLSAGAMLALGSTSLIGCSPVLPNEIPAGVRWEDLR
Mvan_4048|M.vanbaalenii_PYR-1 -----------------------------MGGSHATAQPPPTQAALADLT
Mflv_4651|M.gilvum_PYR-GCK ---------------------------------MTVPTDLRRDEALRSLR
MSMEG_1746|M.smegmatis_MC2_155 -------------------------------------MLQVHPTMVDELA
TH_1540|M.thermoresistible__bu ------------------------------------------------MA
:
Mb0064|M.bovis_AF2122/97 SALGGKVLQPDDGPQFATAKQVFNTNYNGYTPAVIVTPTSQLDVQKAMAF
Rv0063|M.tuberculosis_H37Rv SALGGKVLQPDDGPQFATAKQVFNTNYNGYTPAVIVTPTSQLDVQKAMAF
MAV_0089|M.avium_104 TALGGKVLLPDS-PQFAGAKQVFNTNYNGSTPAAVVTPTSAADVQKAMAF
MMAR_1179|M.marinum_M SSIGGRVLLPSNGASFTSGKQVFNSLYNNSNPAAVVTVTSQADVEKAVAF
MUL_1431|M.ulcerans_Agy99 SSIGGRVLLPSNGASFTSGKQVFNSLYNNSNPAAVVTVTSQADVEKAVAF
MAB_2456|M.abscessus_ATCC_1997 RSLSGALVVRGDAAMDESGR-AFNPLFDVNHPGAVAFCASDQDVARCVEF
Mvan_4048|M.vanbaalenii_PYR-1 RTLRGSVIGPDG-GRFTELATPRNLRFAATMPRAVVLCADADDVAKTVLW
Mflv_4651|M.gilvum_PYR-GCK DQVATPVALPGEPGYERCRP---WNVVAPVEPAAVVLATSPEDVAGTVRF
MSMEG_1746|M.smegmatis_MC2_155 MSMPERVHLPGSPEYGDAVAIWNGAVTER--PAAVVRPTSSGDVRNVVRF
TH_1540|M.thermoresistible__bu ALPALSFRRGAQGYEQARRTTVWNGLLPDRFPDVIVQAVTVDDVVTAVRY
. * .:. . ** : :
Mb0064|M.bovis_AF2122/97 AAANNLKVAPRGGGHSYVGASTANGAMVLDLRQLPGDINYDATTGRVTVT
Rv0063|M.tuberculosis_H37Rv AAANNLKVAPRGGGHSYVGASTANGAMVLDLRQLPGDINYDATTGRVTVT
MAV_0089|M.avium_104 AAAHNLKVAPRSGGHSYTGASTANGTMVLDLRQLPGDANYDAATGQVTVT
MMAR_1179|M.marinum_M AAANKLKIAPRGGGHSYIGASTAVGAMVIDLRGLPGGVNFDAGSGTVTVP
MUL_1431|M.ulcerans_Agy99 AAANKLKIAPRGGGHSYIGASTAVGAMVIDLRGLPGGVNFDAGSGTVTVP
MAB_2456|M.abscessus_ATCC_1997 ATSARLPIAARGGGHSYAAYCIPNDGLVVDLARMAAVS---VTGTRAVVG
Mvan_4048|M.vanbaalenii_PYR-1 ARNTGTSFAIRGGGHNYAEASSSRG-VIISTRQMTAATIDGATLRAHAGV
Mflv_4651|M.gilvum_PYR-GCK AAAHGFRVTVQATGHGAVGVG--PDTILIQTSAMRHVS-VDVVNGTARVG
MSMEG_1746|M.smegmatis_MC2_155 AHEQRIPITVRAGGHDWGGRALNEGGLVIDLSSMRKVY-IDPAAREALVE
TH_1540|M.thermoresistible__bu AADNDLRIGVCSGGHSWAASHLRDGGMLLDVSRLDHCG-IDADRMVAVAG
* . . **. . :::. :
Mb0064|M.bovis_AF2122/97 PATGLYAMHQVLAAAGRGIPTGTCPTVGVAGHALGGGLGANSRHAGLLCD
Rv0063|M.tuberculosis_H37Rv PATGLYAMHQVLAAAGRGIPTGTCPTVGVAGHALGGGLGANSRHAGLLCD
MAV_0089|M.avium_104 PATSLYTMHRTLAAVGRGVPTGTCPSVGAAGHALGGGMGAQSRHAGLLCD
MMAR_1179|M.marinum_M AGVSLTDVHQVLAGAGRAIPTGSCPTVGVGGLALGGGMGADSRHAGLTCD
MUL_1431|M.ulcerans_Agy99 AGVSLTDVHQVLAGAGRAIPTGSCPTVGVGGLALGGGMGADSRHAGLTCN
MAB_2456|M.abscessus_ATCC_1997 AGARLIDVYAGIAGAGRMLAGGSCPTVGIAGLTLGGGVGVLTRRFGLTCD
Mvan_4048|M.vanbaalenii_PYR-1 RNADLAALLPQGGGGRLLLPGGNCPGVGVVGLTLGGGIGPNAPWAGLTAD
Mflv_4651|M.gilvum_PYR-GCK AGARWQDVLDVATPHGLAPLCGSAPGVGVIGYLTGGGIGPLVRTYGLSSD
MSMEG_1746|M.smegmatis_MC2_155 GGATAEDVVQAAERHGLTAAAPNLADVGFTGFTLGGGYGPLNGIAGLGVD
TH_1540|M.thermoresistible__bu PGKIGSELAGELDKLDLFFPAGHCEGICLGGYLLQGGYGWNSRVLGPACE
: : * ** * * :
Mb0064|M.bovis_AF2122/97 QLTSASVVLPSGQAVTASATDHPDLFWALRGGGGGNFGVTTSLTFATFPS
Rv0063|M.tuberculosis_H37Rv QLTSASVVLPSGQAVTASATDHPDLFWALRGGGGGNFGVTTSLTFATFPS
MAV_0089|M.avium_104 QLTSASVVLPSGQAVTASTASNPDLFWALRGGGGGNFGVTTSLTFATFPT
MMAR_1179|M.marinum_M ALRSATVVLPSGETVTASAEDHPDLFWALRGGGGGNFGVTTAMTFETFPV
MUL_1431|M.ulcerans_Agy99 ALRSATVVLPSGETVTASAEDHPDLFWALRGGGGGNFGVTTAMTFETFPV
MAB_2456|M.abscessus_ATCC_1997 QLISARVVTADGKIRVVSADTESDLFWAIRGVGGGNFCIATELAFETAAS
Mvan_4048|M.vanbaalenii_PYR-1 RLRKVTMVTAHGDVVTASPTENPDLFWGLRGGAGGNFGVVTDLEYELVEV
Mflv_4651|M.gilvum_PYR-GCK HVRSFDVVTGGGRLLRAAPDENADLFWGLRG-GKATLGIVTSAEIDLLPV
MSMEG_1746|M.smegmatis_MC2_155 NLLEAHVVLADGRSVTANAADEADLLWALRG-GGANFGVVTHLRVRLHPV
TH_1540|M.thermoresistible__bu SIVGVDVVTADGEVLYCDDETNADLFWAARGSGPGFFAVVTSFHLRLHPR
: :* * ..**:*. ** . . : :.*
Mb0064|M.bovis_AF2122/97 --GDLDVVNLNFPPQSFAQVLVGWQNWLRTADRGS---WALADATVDP--
Rv0063|M.tuberculosis_H37Rv --GDLDVVNLNFPPQSFAQVLVGWQNWLRTADRGS---WALADATVDP--
MAV_0089|M.avium_104 --KDVDAVNLNFPPQSFAQVLVGWQNWLRTADPNS---WALADATVDA--
MMAR_1179|M.marinum_M --ADSDVVRVDFTPSSAAQVLAGWQTWLAAADRDT---WGLVDMSVSA--
MUL_1431|M.ulcerans_Agy99 --ADSDVVRVDFTPSSAAQVLAGWQTWLAAADRDT---WGLVDMSVSA--
MAB_2456|M.abscessus_ATCC_1997 --TDLTVFTLDYAAGEMAPIVHRWLTFMAGAPDEL---WTTLHAIGGA--
Mvan_4048|M.vanbaalenii_PYR-1 PVRRATTAELSVTGADAATRVALAFQQLRADAERI---VTGNLYLGHA--
Mflv_4651|M.gilvum_PYR-GCK P--EFYGGAVYFAADDVADVLRAWAAWCVDLPETINTSIAIQQLPPLP--
MSMEG_1746|M.smegmatis_MC2_155 P--HLATGVLMFPWEQAADVMRRYDALAPSLPDGLT---AQIGVIPAP--
TH_1540|M.thermoresistible__bu P-PVCGSCLYIYPAEAIDDVYTWALTSGRDVDDRVELQIIASRDLPALGL
.
Mb0064|M.bovis_AF2122/97 LGTHCRILATCPAGSGGSVAAAIVSAVGTQPT-GTENHTFNYLDLVRYLA
Rv0063|M.tuberculosis_H37Rv LGTHCRILATCPAGSGGSVAAAIVSAVGTQPT-GTENHTFNYLDLVRYLA
MAV_0089|M.avium_104 MGVHCRILATCPAGSGNSAAAAITQAVGIQPS-GVETHTFNYMDLVNYLA
MMAR_1179|M.marinum_M AQANCHVLATCPAGAGRGVADAIKSAVGAQPT-GIETKTFNHMELVKYLA
MUL_1431|M.ulcerans_Agy99 AQANCHVLATCPAGAGRGVADAIKSAVGAQPT-GIETKTFNHMELVKYLA
MAB_2456|M.abscessus_ATCC_1997 IPQCRIVGCVAQGVNSQDVIESLRGGIGVRAA-DSFIAEMTFLDAMKFMG
Mvan_4048|M.vanbaalenii_PYR-1 AGDVEAALTTQLLVDEADARDLLAPLTAIPGL-TAEITEQPWWGAYAWYV
Mflv_4651|M.gilvum_PYR-GCK EVPEPLAGRMTVAVRYAAVGDVDEAEQALAPMRAVATPVLDTVGVLPYAA
MSMEG_1746|M.smegmatis_MC2_155 DGGPVVFVAPYWVGDPDEGDRWVAELRRLGTPVVDQVAPMSPSAQLRLLD
TH_1540|M.thermoresistible__bu NQPSIVVASPVFAESESAAAQALAVLGDCPAVDRAIIKIPYATTDLAGWY
Mb0064|M.bovis_AF2122/97 VGNLNPSPLGYVGGSD-----------VFTTITPATAQGIASAVDAFPRG
Rv0063|M.tuberculosis_H37Rv VGNLNPSPLGYVGGSD-----------VFTTITPATAQGIASAVDAFPRG
MAV_0089|M.avium_104 VGNLNPQPLGYVGGSD-----------VFPTVNAAVAQGIAAAVNAFPRN
MMAR_1179|M.marinum_M GGSSASSPRGFVAGSD-----------VIGTVNSAAAQAIVAAVGKWPPA
MUL_1431|M.ulcerans_Agy99 GGSSASSPRGFVAGSD-----------VIGTVNSAAAQAIVAAVGKSPPA
MAB_2456|M.abscessus_ATCC_1997 GCTTLTVAQCHPSWTGPGSGQLKREAFVASSRMVPHPDVDTARIETLLAG
Mvan_4048|M.vanbaalenii_PYR-1 TPPSPAYPFWDRSLYAD--------QFLSGDALAAALEVVRRFPAGNDPE
Mflv_4651|M.gilvum_PYR-GCK IGAVHADPVDPMPINED---------QALLRELPPEAVEALLAAAGPESG
MSMEG_1746|M.smegmatis_MC2_155 PLVPRGRHYEMR---------------TVNVDTLSPGVIDALVAAGSSRT
TH_1540|M.thermoresistible__bu GAIMTNYPTGHRYATDN----------MWTSASPEALLPGIHRIIETLPP
Mb0064|M.bovis_AF2122/97 AGRMLAIMHALDGALATVSPGATAFPWRRQSALVQWYVET---------S
Rv0063|M.tuberculosis_H37Rv AGRMLAIMHALDGALATVSPGATAFPWRRQSALVQWYVET---------S
MAV_0089|M.avium_104 AGRMLAIMHALDGALATVAPAATAFPWRRQSALVQWYVET---------S
MMAR_1179|M.marinum_M SGRASALVDTLTGAVGDIDPTATAFPWRRQSAVVQWYVETP-------SS
MUL_1431|M.ulcerans_Agy99 SGWASALVDTLTGAVGDIDPTATAFPWRRQSAVVQWYVETP-------SS
MAB_2456|M.abscessus_ATCC_1997 KPGLTFIFDSLGGAVRRISPDATAFPHRQAAACIQIYHGVG---------
Mvan_4048|M.vanbaalenii_PYR-1 RYGALGLYGWVGGAVNDVAPDATAYVHRTARILVEMSSGWSPAPSGAPVA
Mflv_4651|M.gilvum_PYR-GCK SPQAIVELRHLGGALAREATHRSAFCHRNAAFSLAVIGVPAP--------
MSMEG_1746|M.smegmatis_MC2_155 SPASVLALHHFHGASTRVGLTDTAFGLRTPHFLVEIVAAWDAD-----SD
TH_1540|M.thermoresistible__bu APSHFLMFNWSP-----TSTRDNLFYGLEADFNLALYAVWN---------
. : :
Mb0064|M.bovis_AF2122/97 GSPSEATSWLNTAHQAVRAYS-VGGYVNYLEVNQP--PARYFGPN-LSRL
Rv0063|M.tuberculosis_H37Rv GSPSEATSWLNTAHQAVRAYS-VGGYVNYLEVNQP--PARYFGPN-LSRL
MAV_0089|M.avium_104 GDPAAATNWLASAHQAVQQYS-VGGYVNYLEANQS--PARYFGPN-LSRL
MMAR_1179|M.marinum_M GQTAAATKWINSAHQAVQQFS-VGGYVNYLEANTA--PSRYFGSN-LSRL
MUL_1431|M.ulcerans_Agy99 GQTAAATKWINSAHQAVQQFS-VGGYVNYLEANTA--LSRYFGSN-LSRL
MAB_2456|M.abscessus_ATCC_1997 ADPAVAHERVSQARDGLGDICGPAAYVNYIDPGMPDWATAYYGDN-LPRL
Mvan_4048|M.vanbaalenii_PYR-1 PIPPDIRDWEDELWETVLPHTTGRSYQNFPDPELADWPRAYYGAN-LDRL
Mflv_4651|M.gilvum_PYR-GCK EIADMVIAHAGAVTGAVAPWSTGGQMPNFAPSYDPARPSRVYTDDTLHWL
MSMEG_1746|M.smegmatis_MC2_155 DDGERHRRWAQSTFDALGAHALPGGYPNLIGPDQRNQADTAYGPN-AERL
TH_1540|M.thermoresistible__bu -DPEDDERFSAWPGTNMAALAHLAMGVGLADENLGRRPARFATDANMARL
: . *
Mb0064|M.bovis_AF2122/97 SAVRQKYDPSRVMFSGLNF------
Rv0063|M.tuberculosis_H37Rv SAVRQKYDPSRVMFSGLNF------
MAV_0089|M.avium_104 SAVRQKYDPGRVMFSGLNY------
MMAR_1179|M.marinum_M TQIRQRYDPSRLMYSGLNF------
MUL_1431|M.ulcerans_Agy99 TQIRQRYDPSRLMYSGLNF------
MAB_2456|M.abscessus_ATCC_1997 RGIAAAYDPKGIFRFAQAVRP----
Mvan_4048|M.vanbaalenii_PYR-1 TRVKATWDPEDVFRYPQGIPPR---
Mflv_4651|M.gilvum_PYR-GCK AGLADRYDPARTLATGQVIRTVV--
MSMEG_1746|M.smegmatis_MC2_155 LAAKDRWDAGNVFTAIPLPSRSSTS
TH_1540|M.thermoresistible__bu DGIRARRDPAGRFHGWMGRV-----
*. :