For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNYSGFTAQPTSSTRRPDAARNAGECRGRPLFEVVEASIAELRAALESGETSSAELTRAYLERIDAYDKS GPTLNALVVINPDAMLEARASDARRGRGELLGPLDGIPYTAKDSYLVKGLTAAAGSPAFEHLVAQRDAFT IERLRAGGAVLIGLTNMPPMANGGMQRGVYGRAESPYNGDFLTAAFGSGSSNGSGTATAASFAAFGLGEE TWSSGRSPASNNALCAYTPSRGVISVRGNWPLVPTMDVVVPHTRSMADMFEVLDVIVADDAETRGDFWRM QSWVEIPAASTVRPASYPTLAPASAAKARAALRGKRFGVPRMYVNADPEAGTGESPGIGGSTGQPIVTRS SVMDLWRVAKLDLERAGAEVILVDFPVVSNYEGDRPGAPTIGTRGLVSPEFLRTETGLLAAWSWDDFLRA NDDPALNRLADVNGEMISPHPAGALPDRYAGFDDDIAEYPACIRENAVSGLGAIPHLPEGLRGLEETRRL DLEEWMDGHRLDAVIFPASADVGAADMDVDQESADRGWRNGVWVANGNLVPRHLGIPTVTVPMGTMSDIG MPVGLTFAGRSYDDAALLTLAAAFEATGMRRTVPPRTPRLG
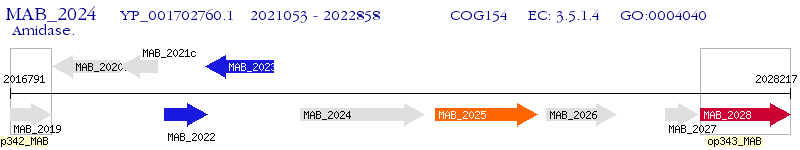
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2024 | - | - | 100% (601) | amidase |
| M. abscessus ATCC 19977 | MAB_3341c | gatA | 2e-23 | 28.39% (310) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. abscessus ATCC 19977 | MAB_1674c | - | 4e-20 | 34.96% (226) | amidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3200 | - | 9e-29 | 38.73% (204) | amidase |
| M. gilvum PYR-GCK | Mflv_4246 | gatA | 3e-22 | 28.61% (346) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. tuberculosis H37Rv | Rv3175 | - | 9e-29 | 38.73% (204) | amidase |
| M. leprae Br4923 | MLBr_01702 | gatA | 2e-22 | 29.25% (294) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. marinum M | MMAR_0937 | gatA_1 | 4e-41 | 28.89% (585) | peptide amidase, GatA_1 |
| M. avium 104 | MAV_3859 | gatA | 9e-22 | 28.32% (346) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. smegmatis MC2 155 | MSMEG_3400 | - | 4e-34 | 39.00% (241) | glutamyl-tRNA(Gln) amidotransferase subunit A |
| M. thermoresistible (build 8) | TH_2639 | - | 7e-23 | 28.85% (357) | PUTATIVE glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. ulcerans Agy99 | MUL_0689 | gatA_1 | 9e-39 | 28.62% (587) | peptide amidase, GatA_1 |
| M. vanbaalenii PYR-1 | Mvan_2115 | gatA | 7e-24 | 28.21% (351) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 MVSHSKDSAETPTLAESVYQRIRRNILEGRIMPGTRVSIRSLAESVGVST
Mb3200|M.bovis_AF2122/97 --------------------------------------------------
Rv3175|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2024|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 MPTREALKRLSFEGLVEYDRRAVTISTLSPKAIRELFTIRLRLELLATEW
Mb3200|M.bovis_AF2122/97 --------------------------------------------------
Rv3175|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2024|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 ALPRLDAETTVALRSVLDRMIVPGISAITWRELNREFHQTFYSCGDSRYL
Mb3200|M.bovis_AF2122/97 --------------------------------------------------
Rv3175|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2024|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4246|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01702|M.leprae_Br4923 --------------------------------------------------
MAV_3859|M.avium_104 --------------------------------------------------
TH_2639|M.thermoresistible__bu --------------------------------------------------
MMAR_0937|M.marinum_M --------------------------------------------------
MUL_0689|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3400|M.smegmatis_MC2_155 LELIHNIWDRIQPYMAIYASAMHDFTEADRQHEHMYQLMLARDLDGLLDA
Mb3200|M.bovis_AF2122/97 --------------------------------------------------
Rv3175|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2024|M.abscessus_ATCC_1997 ----------------------------------------------MNYS
Mflv_4246|M.gilvum_PYR-GCK --------------------------MSELIKLDAATLADKISSKEVSSA
Mvan_2115|M.vanbaalenii_PYR-1 --------------------------MTDLIKLDAATLAAKIAAREVSAT
MLBr_01702|M.leprae_Br4923 ----------------------MIGSLHEIIRLDASTLAAKIVAKELSSV
MAV_3859|M.avium_104 --------------------------MNEIIRSDAATLAARIAAKELSSV
TH_2639|M.thermoresistible__bu --------------------MSQGKSTDDLIRLDAATLGAKIAAKEISST
MMAR_0937|M.marinum_M ---------------------------MELPEFTIAETQTAFERGEWTAA
MUL_0689|M.ulcerans_Agy99 ---------------------------MELPEFTIAETQTAFERGEWTAA
MSMEG_3400|M.smegmatis_MC2_155 TTRHLEHTAEAIVSALGADTTTGRGNNVNYEELSISSFHRLVESGGATSA
Mb3200|M.bovis_AF2122/97 -------------------MAMSAKASDDIAWLPATAQLAVLAAKKVSSA
Rv3175|M.tuberculosis_H37Rv -------------------MAMSAKASDDIAWLPATAQLAVLAAKKVSSA
MAB_2024|M.abscessus_ATCC_1997 GFTAQPTSSTRRPDAARNAGECRGRPLFEVVEASIAELRAALESGETSSA
: : . ::.
Mflv_4246|M.gilvum_PYR-GCK EVTQAFLDQIAATD------GDYHAFLHVGAEQALAAAQTVDR----AVA
Mvan_2115|M.vanbaalenii_PYR-1 EVTQACLDQIAATD------AEYHAFLHVAGDQALAAAATVDKAIHEATA
MLBr_01702|M.leprae_Br4923 EITQACLDQIEATD------DTYRAFLHVAAEKALSAAAAVDK----AVA
MAV_3859|M.avium_104 EATQACLDQIAATD------ERYHAFLHVAADEALGAAAVVDE----AVA
TH_2639|M.thermoresistible__bu ELTQACLDRIAATD------DRYNAFLHVAANKALSAAARVDA----AVA
MMAR_0937|M.marinum_M GLTDCYLRRIREID----QSGPMLRSIIEVNPDALAIAEALDA----ERS
MUL_0689|M.ulcerans_Agy99 GLTDCYLRRIREID----QSGPMLRSIIEVNSDALAIAEALDA----ERS
MSMEG_3400|M.smegmatis_MC2_155 DLTAWYLDRIATLDSIDNPDGPQLNSVVTVNPAALDEARALDE----KYA
Mb3200|M.bovis_AF2122/97 ELVELYLSRIDTYN-------ASLNAIVTVDPDAARRVAKRSD----AAR
Rv3175|M.tuberculosis_H37Rv ELVELYLSRIDTYN-------ASLNAIVTVDPDAARRVAKRSD----AAR
MAB_2024|M.abscessus_ATCC_1997 ELTRAYLERIDAYD----KSGPTLNALVVINPDAMLEARASDAR-----R
. * :* : : * . .
Mflv_4246|M.gilvum_PYR-GCK AGEHLPSPLAGVPLALKDVFTT-TDMPTTCGSKVLEGWTSPYDATVTAKL
Mvan_2115|M.vanbaalenii_PYR-1 AGERLPSPLAGVPLALKDVFTT-TDMPTTCGSKILEGWTSPYDATVTAKL
MLBr_01702|M.leprae_Br4923 AGGQLSSTLAGVPLALKDVFTT-VDMPTTCGSKILQGWHSPYDATVTTRL
MAV_3859|M.avium_104 AGERLPSPLAGVPLALKDVFTT-VDMPTTCGSKILQGWRSPYDATVTTKL
TH_2639|M.thermoresistible__bu AGEELPSPLAGVPLALKDVFTT-IDMPTTCGSKILDGWQAPYDATVTVAL
MMAR_0937|M.marinum_M -GGRIRGALHGVPVVIKDSIDTGDKMATTAGSLALEGNIATRDAFVVKQL
MUL_0689|M.ulcerans_Agy99 -GGRIRGALHGVPVVIKDSIDTGDKMATTAGSLALEGNIATRDAFVVKQL
MSMEG_3400|M.smegmatis_MC2_155 RTGTLVGPLHGVPILIKDQGET-KGIPTAFGSTAFADYIPDTDATVVDRL
Mb3200|M.bovis_AF2122/97 ARGDELGPLHGLPITVKDSYET-AGMRTTCGRRDLADYVPTQDAEAVARL
Rv3175|M.tuberculosis_H37Rv ARGDELGPLHGLPITVKDSYET-AGMRTTCGRRDLADYVPTQDAEAVARL
MAB_2024|M.abscessus_ATCC_1997 GRGELLGPLDGIPYTAKDSYLV-KGLTAAAGSPAFEHLVAQRDAFTIERL
..* *:* ** . : :: * : . ** . *
Mflv_4246|M.gilvum_PYR-GCK RSAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWD----
Mvan_2115|M.vanbaalenii_PYR-1 RAAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWN----
MLBr_01702|M.leprae_Br4923 RAAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWN----
MAV_3859|M.avium_104 RAAGIPILGKTNMDEFAMG-----------SSTENSAYGPTRNPWN----
TH_2639|M.thermoresistible__bu RAAGIPILGKTNLDEFAMG-----------SSTENSAFGPTRNPWD----
MMAR_0937|M.marinum_M RDAGAVILGKANMSEWG------YMRSTRPCSGWSSRGGQVRNPYV----
MUL_0689|M.ulcerans_Agy99 RDAGAVILGKANMSEWGNMSEWGYMRSTRPCSGWSSRGGQVRNPYV----
MSMEG_3400|M.smegmatis_MC2_155 RKAGAVILGKTAMCDFAAG-----------WFSFSSRTDHTKNPYD----
Mb3200|M.bovis_AF2122/97 RRAGAIIMGKTNMPTGNQD-----------VQASNPVFGRTNNPWD----
Rv3175|M.tuberculosis_H37Rv RRAGAIIMGKTNMPTGNQD-----------VQASNPVFGRTNNPWD----
MAB_2024|M.abscessus_ATCC_1997 RAGGAVLIGLTNMPPMANG------------GMQRGVYGRAESPYNGDFL
* .* ::* : : . ...*:
Mflv_4246|M.gilvum_PYR-GCK TERVPGGSGGGSAAALAAFQAPLAIGTDTGGSIRQPAALTATVGVKPTYG
Mvan_2115|M.vanbaalenii_PYR-1 VDCVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYG
MLBr_01702|M.leprae_Br4923 VDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYG
MAV_3859|M.avium_104 VERVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYG
TH_2639|M.thermoresistible__bu VNRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPAALTATVGVKPTYG
MMAR_0937|M.marinum_M LDRSPLGSSSGSAVAVAANLCVAALGAEVDGSIVRPASSNSIVGLKPTVG
MUL_0689|M.ulcerans_Agy99 LDRSPLGSSSSSAVAVAANLCVAALGAEVDGSIVRPASSNSIVGLKPTVG
MSMEG_3400|M.smegmatis_MC2_155 LDRETGGSSAGTAAAVTANLCLVGIGEDTGGSIRLPSSFTNLFGLRVTTG
Mb3200|M.bovis_AF2122/97 AARTSGGSAGGGAAATAAGLTSFDYGSEIGGSTRIPAHYCGLYGHKSTWR
Rv3175|M.tuberculosis_H37Rv AARTSGGSAGGGAAATAAGLTSFDYGSEIGGSTRIPAHYCGLYGHKSTWR
MAB_2024|M.abscessus_ATCC_1997 TAAFGSGSSNGSGTATAASFAAFGLGEETWSSGRSPASNNALCAYTPSRG
**. . ..* :* * : .* *: . :
Mflv_4246|M.gilvum_PYR-GCK TVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHDPLDS--------
Mvan_2115|M.vanbaalenii_PYR-1 TVSRYGLIACASSLDQGGPCARTVLDTALLHQVIAGHDPRDS--------
MLBr_01702|M.leprae_Br4923 TVSRYGLVACASSLDQGGPCARTVLDTALLHAVIAGHDARDS--------
MAV_3859|M.avium_104 TVSRYGLVACASSLDQGGPCARTVLDTALLHQVIAGHDIRDS--------
TH_2639|M.thermoresistible__bu TVSRYGLVACASSLDQGGPCGRTVLDTALLHQVIAGHDAKDS--------
MMAR_0937|M.marinum_M LLSRSGVIGVASPQDMVGPMARTVTDVATLLTVMTGVDDSD---------
MUL_0689|M.ulcerans_Agy99 LLSRSGVIGVASPQDMVGPMARTVTDVATLLTVMTGVDDSD---------
MSMEG_3400|M.smegmatis_MC2_155 LVSRTGFSPLVHFQDTPGPMARNVSDLAAVLDVIVGYDPTDS--------
Mb3200|M.bovis_AF2122/97 SVPLVGHIPSAP--GNPGRWG-QADMACAGVQVRGARDIIPA--------
Rv3175|M.tuberculosis_H37Rv SVPLVGHIPSAP--GNPGRWG-QADMACAGVQVRGARDIIPA--------
MAB_2024|M.abscessus_ATCC_1997 VISVRGNWPLVPTMDVVVPHTRSMADMFEVLDVIVADDAETRGDFWRMQS
:. * . . * . *
Mflv_4246|M.gilvum_PYR-GCK ----TSVEAPVPDVVAAARTGAG----GDLTGVRIGVVKQLRSGEGYQAG
Mvan_2115|M.vanbaalenii_PYR-1 ----TSVNAAVPDVVGAARAGAR----GDLKGVRIGVVKQLRSGEGYQPG
MLBr_01702|M.leprae_Br4923 ----TSVEAAVPDIVGAAKAGES----GDLHGVRVGVVRQLR-GEGYQSG
MAV_3859|M.avium_104 ----TSVDAPVPDVVGAARAGAA----GDLKGVRVGVVKQLR-GEGYQPG
TH_2639|M.thermoresistible__bu ----TSLTAPVPDVVAAARAGAS----GDLKGVRVGVVRQLR-GEGIQPG
MMAR_0937|M.marinum_M ----PTTRAGGAHTATDYRRFLD---PAALQGARLGVARERFGA---HEA
MUL_0689|M.ulcerans_Agy99 ----PTTRAGGAHTATDYRRFLD---PAALQGARLGVARERFGA---HEA
MSMEG_3400|M.smegmatis_MC2_155 ----YTALATSGPEVGDYGEALDGVDADTLSGFRVGVLTDAFGAGDDQEL
Mb3200|M.bovis_AF2122/97 ----LEATVGPMRADGGFSYALAPPRAGALKDFRVAVWAEDP-HCPIDAD
Rv3175|M.tuberculosis_H37Rv ----LEATVGPMRADGGFSYALAPPRAGALKDFRVAVWAEDP-HCPIDAD
MAB_2024|M.abscessus_ATCC_1997 WVEIPAASTVRPASYPTLAPASAAKARAALRGKRFGVPRMYVNADPEAGT
. * . *..*
Mflv_4246|M.gilvum_PYR-GCK VLAS-FNAAVDQLTALGAEVTE-VDCPHFDYSLPAYYLILP---------
Mvan_2115|M.vanbaalenii_PYR-1 VLAS-FTAAVEQLTALGAEVSE-VDCPHFDHSLAAYYLILP---------
MLBr_01702|M.leprae_Br4923 VLAS-FQAAVEQLIALGATVSE-VDCPHFDYALAAYYLILP---------
MAV_3859|M.avium_104 VLAS-FEAAVEQLTALGAEVSE-VDCPHFEYALAAYYLILP---------
TH_2639|M.thermoresistible__bu VLAS-FDAAVEQLAALGAEVIE-VDCPNIDHALAAYYLIMP---------
MMAR_0937|M.marinum_M TDAL-IEGALGQLAALGAEIVDPIQASSLPFFGDLELELFR---------
MUL_0689|M.ulcerans_Agy99 TDAL-IEGALGQLAALGAEIVDPIQASSLPFFGDLELELFR---------
MSMEG_3400|M.smegmatis_MC2_155 TNSV-VRAAIDRLRHHGTGVVDGIVLGNLETW-VAETSLYT---------
Mb3200|M.bovis_AF2122/97 VRRA-MDDAVAALRAAGAHVVE--QPATIPVDMAVSHNIFQ---------
Rv3175|M.tuberculosis_H37Rv VRRA-MDDAVAALRAAGAHVVE--QPATIPVDMAVSHNIFQ---------
MAB_2024|M.abscessus_ATCC_1997 GESPGIGGSTGQPIVTRSSVMDLWRVAKLDLERAGAEVILVDFPVVSNYE
. : : : : : :
Mflv_4246|M.gilvum_PYR-GCK -------------------SEVSSNLAKFDGMRYGLRAGDDGTHSAEEVM
Mvan_2115|M.vanbaalenii_PYR-1 -------------------SEVSSNLAKFDGMRYGLRVGDDGTTSAEEVM
MLBr_01702|M.leprae_Br4923 -------------------SEVSSNLARFDAMRYGLRVGDDGSRSAEEVI
MAV_3859|M.avium_104 -------------------SEVSSNLARFDAMRYGLRIGDDGSHSAEEVM
TH_2639|M.thermoresistible__bu -------------------SEVSSNLARFDAMRYGLRVGDDGTRSAEEVM
MMAR_0937|M.marinum_M -------------------YGLKASLNGYLGAHPRAAVGSLDELIAFNRA
MUL_0689|M.ulcerans_Agy99 -------------------YGLKASLNGYLGAHPRAAVGSLDEPIAFNRA
MSMEG_3400|M.smegmatis_MC2_155 -------------------IQSKFDLEKFLGSRRHTGPTTIREIVDHGDF
Mb3200|M.bovis_AF2122/97 -------------------SLVFG---AFAVDRSTLSPASAAALGLRAVR
Rv3175|M.tuberculosis_H37Rv -------------------SLVFG---AFAVDRSTLSPASAAALGLRAVR
MAB_2024|M.abscessus_ATCC_1997 GDRPGAPTIGTRGLVSPEFLRTETGLLAAWSWDDFLRANDDPALNRLADV
Mflv_4246|M.gilvum_PYR-GCK ALTRAAGFGPEVKRRIMIGAYALSAGYYDAYYN----------------Q
Mvan_2115|M.vanbaalenii_PYR-1 AMTRAAGFGAEVKRRIMIGTYALSAGYYDAYYN----------------Q
MLBr_01702|M.leprae_Br4923 ALTRAAGFGPEVKRRIMIGAYALSAGYYDSYYS----------------Q
MAV_3859|M.avium_104 ALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYN----------------Q
TH_2639|M.thermoresistible__bu ALTRAAGFGPEVKRRIMIGTYALSAGYYDAYYN----------------Q
MMAR_0937|M.marinum_M HAGQVMPYFGQEFLEQSQAKGDLTDPQYLRVRA----------------E
MUL_0689|M.ulcerans_Agy99 HAGQVMPYFGQEFLEQSQAKGDLTDSQYLRVRA----------------E
MSMEG_3400|M.smegmatis_MC2_155 HP---LTDLLADIADGPSNPEDHPEYFKKRTRQ----------------E
Mb3200|M.bovis_AF2122/97 HPRGEAANALGATLQSHR-AWLFADAARHEMRD----------------R
Rv3175|M.tuberculosis_H37Rv HPRGEAANALGATLQSHR-AWLFADAARHEMRD----------------R
MAB_2024|M.abscessus_ATCC_1997 NGEMISPHPAGALPDRYAGFDDDIAEYPACIRENAVSGLGAIPHLPEGLR
.
Mflv_4246|M.gilvum_PYR-GCK AQKVRTLIARDLDAAYQKVDVLVSPATPTTAFRLGEKVDDPLSMYLFD--
Mvan_2115|M.vanbaalenii_PYR-1 AQKVRTLIARDLDEAYRSVDVLVSPATPSTAFRLGEKVDDPLAMYLFD--
MLBr_01702|M.leprae_Br4923 AQKVRTLIARDLDKAYRSVDVLVSPATPTTAFSLGEKADDPLAMYLFD--
MAV_3859|M.avium_104 AQKVRTLIARDLDAAYESVDVVVSPATPTTAFGLGEKVDDPLAMYLFD--
TH_2639|M.thermoresistible__bu AQKVRTLVARDFDAAYEKVDVLVSPTTPTTAFPLGEKVDHPLAMYLFD--
MMAR_0937|M.marinum_M LRRLAGADGIDKALREHRLDAIVAPTEGSPAFAIDPVVGDNILPGGCS--
MUL_0689|M.ulcerans_Agy99 LRRLAGADGIDKALREHRLDAIVAPTEGSPAFAIDPVVGDNILPGGCS--
MSMEG_3400|M.smegmatis_MC2_155 DWRRT----LLAKMADAEIDFLVYPTVQVPAPTRADLAAKRWTALNFP--
Mb3200|M.bovis_AF2122/97 WAGFFNEFDVLLLPVTPTPAPLHHNKDHDRLGRTIDVDGVSRSYWDQL--
Rv3175|M.tuberculosis_H37Rv WAGFFNEFDVLLLPVTPTPAPLHHNKDHDRLGRTIDVDGVSRSYWDQL--
MAB_2024|M.abscessus_ATCC_1997 GLEETRRLDLEEWMDGHRLDAVIFPASADVGAADMDVDQESADRGWRNGV
:
Mflv_4246|M.gilvum_PYR-GCK ---LCTLPLNLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRVGA
Mvan_2115|M.vanbaalenii_PYR-1 ---LCTLPLNLAGHCGMSVPSGLSADDNLPVGLQIMAPALADDRLYRVGA
MLBr_01702|M.leprae_Br4923 ---LCTLPLNLAGHCGMSVPSGLSSDDDLPVGLQIMAPALADDRLYRVGA
MAV_3859|M.avium_104 ---LCTLPLNLAGHCGMSVPSGLSPDDDLPVGLQIMAPALADDRLYRVGA
TH_2639|M.thermoresistible__bu ---LCTLPLNLAGHCGMSVPSGLSPDDNLPVGLQIMAPALADDRLYRVGA
MMAR_0937|M.marinum_M ---TPPAVAG---YPHICVPAGYFCG--LPVGLSLFAGAFQEPKLIGYAY
MUL_0689|M.ulcerans_Agy99 ---TPPAVAG---YPHICVPAGYFCG--LPVGLSLFAGAFQEPKLIGYAY
MSMEG_3400|M.smegmatis_MC2_155 ---TNTVIASQTSLPAMSIPVGFTDSG-LPVGLEVVGRPLTERALLRFAR
Mb3200|M.bovis_AF2122/97 ---KWNALANIAGTPATTMPITTTATG-LPIGIQAMGPAGGDRTTVEFAA
Rv3175|M.tuberculosis_H37Rv ---KWNALANIAGTPATTMPITTTATG-LPIGIQAMGPAGGDRTTVEFAA
MAB_2024|M.abscessus_ATCC_1997 WVANGNLVPRHLGIPTVTVPMGTMSDIGMPVGLTFAGRSYDDAALLTLAA
:* :*:*: . . : .
Mflv_4246|M.gilvum_PYR-GCK AYEAARGPLPTAL----------
Mvan_2115|M.vanbaalenii_PYR-1 AYEAARGPLPTAL----------
MLBr_01702|M.leprae_Br4923 AYEAARGPLPSAI----------
MAV_3859|M.avium_104 AYEAARGPLRSAI----------
TH_2639|M.thermoresistible__bu AYEAARGPLPAAV----------
MMAR_0937|M.marinum_M AFEQATGVRRPPRFVPTLST---
MUL_0689|M.ulcerans_Agy99 AFEQATGVRRPPRFVPTLST---
MSMEG_3400|M.smegmatis_MC2_155 AWELAEAPRRQPQIEASKARVTM
Mb3200|M.bovis_AF2122/97 LLTEVLGGFRVPPL---------
Rv3175|M.tuberculosis_H37Rv LLTEVLGGFRVPPL---------
MAB_2024|M.abscessus_ATCC_1997 AFEATGMRRTVPPRTPRLG----
. .