For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MEFLQKMRGATARLAAVAAVAAAILPGLIGVTGGSAIAGAFSRPGLPVEYLQVPSPAMGRDIVVEFQPGG PHAVYLLDGQRAREDYNGWDIETTAFEDYYQSGISVVMPVGGQSSNYTDWYNPAKGKDGVWTYKWETFLT TELPQWLGVNKGISPAANAVVGLSMAGPSALTLAIYHPQQFVYAGALSAPLHPSDQKWQIRVAMSDAGGF NADDMWGPDSDPAWARNDPFLHIDQLIANNTRLWIYCGSGDATDLDKDRNGLEVISGGVIEGQVIDSDKK FAEAYGSAGGANAHFEFPKGGIHNWTYWGNQLRAMKADLVGYLTKPGTAAV
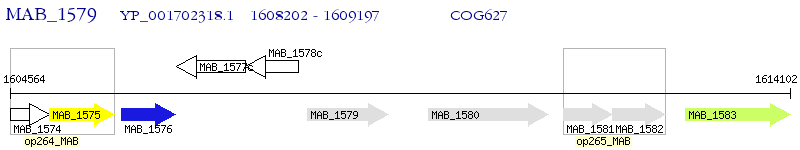
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1579 | - | - | 100% (331) | antigen 85-B precursor |
| M. abscessus ATCC 19977 | MAB_0176 | - | e-141 | 72.12% (330) | antigen 85-A precursor |
| M. abscessus ATCC 19977 | MAB_0177 | - | e-139 | 71.83% (323) | antigen 85-A/B/C precursor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1918c | fbpB | 1e-111 | 61.78% (314) | secreted antigen 85-B fbpB (85B) (antigen 85 complex B) |
| M. gilvum PYR-GCK | Mflv_3335 | - | 1e-116 | 59.58% (334) | putative esterase |
| M. tuberculosis H37Rv | Rv1886c | fbpB | 1e-111 | 61.78% (314) | secreted antigen 85-B fbpB (85B) (antigen 85 complex B) |
| M. leprae Br4923 | MLBr_00097 | fbpA | 1e-107 | 60.00% (330) | antigen 85A, mycolytransferase |
| M. marinum M | MMAR_0328 | fbpC | 1e-113 | 57.01% (335) | secreted antigen 85-C FbpC |
| M. avium 104 | MAV_5183 | - | 1e-110 | 55.82% (335) | antigen 85-C |
| M. smegmatis MC2 155 | MSMEG_3580 | - | 1e-113 | 60.50% (319) | antigen 85-C |
| M. thermoresistible (build 8) | TH_2078 | fbpA | 1e-103 | 56.71% (328) | SECRETED ANTIGEN 85-A FBPA (MYCOLYL TRANSFERASE 85A) |
| M. ulcerans Agy99 | MUL_4793 | fbpC | 1e-113 | 57.01% (335) | secreted antigen 85-C FbpC |
| M. vanbaalenii PYR-1 | Mvan_3061 | - | 1e-115 | 61.11% (324) | putative esterase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3335|M.gilvum_PYR-GCK MKWKEKARG---TRKSLVRRVAAAALAAAALPGLIGVAGGSATAGAFSRP
Mvan_3061|M.vanbaalenii_PYR-1 MKLKEKWRG---TARAAMRRVAAAALAAVALPGLIGFAGGSATAGAFSRP
MSMEG_3580|M.smegmatis_MC2_155 MRGIAAWK--------ALRRFVIGALAALMLPGLIGFAGGSASAGAFSRP
MMAR_0328|M.marinum_M MKFVEKLR---GAAAGAPRRLTIAAIGAALLSGLVGAVGGAATAGAFSRP
MUL_4793|M.ulcerans_Agy99 MKFVEKLR---GAAAGAPRRLTIAAIGAALLSGLVGAVGGAATAGAFSRP
MAV_5183|M.avium_104 MSFIEKVRKLRGAAATMPRRLAIAAVGASLLSGVAVAAGGSPVAGAFSKP
TH_2078|M.thermoresistible__bu MKLVAKLRG---AARALPRRLTVAALAAALLPGLVGAFGGSATAGAFSRP
Mb1918c|M.bovis_AF2122/97 MTDVSRKIR---AWG---RRLMIGTAAAVVLPGLVGLAGGAATAGAFSRP
Rv1886c|M.tuberculosis_H37Rv MTDVSRKIR---AWG---RRLMIGTAAAVVLPGLVGLAGGAATAGAFSRP
MLBr_00097|M.leprae_Br4923 MKFVDRFRG---AVAGMLRRLVVEAMGVALLSALIGVVG-SAPAEAFSRP
MAB_1579|M.abscessus_ATCC_1997 MEFLQKMRG------ATARLAAVAAVAAAILPGLIGVTGGSAIAGAFSRP
* * : .. *..: * :. * ***:*
Mflv_3335|M.gilvum_PYR-GCK GLPVEYLDVFSPSMNRDIRIQFQGGG--PHAVYLLDGLRAQEDYNGWDIN
Mvan_3061|M.vanbaalenii_PYR-1 GLPVEYLDVFSPSMGRDIRVQFQGGG--PRAVYLLDGLRAQEDYNGWDIN
MSMEG_3580|M.smegmatis_MC2_155 GLPVEYLDVFSPSMNRDIRVQFQGGG--PHAVYLLDGLRAQDDYNGWDIN
MMAR_0328|M.marinum_M GLPVEYLQIPSAAMGRDIKVQFQGGG--AHSVYLLDGLRAQDDYNGWDIN
MUL_4793|M.ulcerans_Agy99 GLPVEYLQIPSAAMGRDIKVQFQGGG--AHSVYLLDGLRAQDDYNGWDIN
MAV_5183|M.avium_104 GLPVEYLEVPSPSMGRNIKVQFQGGG--PHAVYLLDGLRAQDDYNGWDIN
TH_2078|M.thermoresistible__bu GLPVEYLMVPSAAMGRDIKVQFQSGGEGSPGVYLLDGLRARDDFNGWDIE
Mb1918c|M.bovis_AF2122/97 GLPVEYLQVPSPSMGRDIKVQFQSGGNNSPAVYLLDGLRAQDDYNGWDIN
Rv1886c|M.tuberculosis_H37Rv GLPVEYLQVPSPSMGRDIKVQFQSGGNNSPAVYLLDGLRAQDDYNGWDIN
MLBr_00097|M.leprae_Br4923 GLPVEYLQVPSPSMGRDIKVQFQNGGANSPALYLLDGLRAQDDFSGWDIN
MAB_1579|M.abscessus_ATCC_1997 GLPVEYLQVPSPAMGRDIVVEFQPGG--PHAVYLLDGQRAREDYNGWDIE
******* : *.:*.*:* ::** ** . .:***** **::*:.****:
Mflv_3335|M.gilvum_PYR-GCK TPAFEWFYQSGLSTIMPVGGQSSFYSDWYQPSRGNGQNYTYKWETFLTQE
Mvan_3061|M.vanbaalenii_PYR-1 TPAFEWFYQSGLSTIMPVGGQSSFYTDWYQPSRGNGQNYTYKWETFLSQE
MSMEG_3580|M.smegmatis_MC2_155 TPAFEWFYQSGLSTIMPVGGQSSFYTDWYQPSKGNGQDYTYKWETFLTQE
MMAR_0328|M.marinum_M TPAFEEYYNSGLSVVMPVGGQSSFYSDWYQPSQGNGQGYTYKWETFLTRE
MUL_4793|M.ulcerans_Agy99 TPAFEEYYNSGLSVVMPVGGQSSFYSDWYQPSQGNGQGYTYKWETFLTRE
MAV_5183|M.avium_104 TPAFEEFYQSGLSVIMPVGGQSSFYSNWYQPSSGNGQNYTYKWETFLTQE
TH_2078|M.thermoresistible__bu TAAFEWYYESGLSVIMPVGGQSSFYSDWYSPACGNNGCQTYKWETFLTQE
Mb1918c|M.bovis_AF2122/97 TPAFEWYYQSGLSIVMPVGGQSSFYSDWYSPACGKAGCQTYKWETFLTSE
Rv1886c|M.tuberculosis_H37Rv TPAFEWYYQSGLSIVMPVGGQSSFYSDWYSPACGKAGCQTYKWETFLTSE
MLBr_00097|M.leprae_Br4923 TTAFEWYYQSGISVVMPVGGQSSFYSDWYSPACGKAGCQTYKWETFLTSE
MAB_1579|M.abscessus_ATCC_1997 TTAFEDYYQSGISVVMPVGGQSSNYTDWYNPAKGKDGVWTYKWETFLTTE
*.*** :*:**:* :******** *::**.*: *: ********: *
Mflv_3335|M.gilvum_PYR-GCK LPAWLEANRGVSRTGNAVVGLSMAGSASLTYAIHHPQQFIYAASLSGFLN
Mvan_3061|M.vanbaalenii_PYR-1 LPAWLEANRGVSRTGNAVVGLSMAGSAALTYAIYHPQQFIYAASLSGFLN
MSMEG_3580|M.smegmatis_MC2_155 LPAWLEANRGVSRTGNAVVGLSMAGSAALTYAIYHPEQFIYASTLSGFLN
MMAR_0328|M.marinum_M MPAWLQANKGVSPTGNAAVGLSMSGGSALVLAAYYPQQFPYAASLSGFLN
MUL_4793|M.ulcerans_Agy99 MPAWLQANKGVSPTGNAAVGLSMSGGSALVLAAYYPQQFPYAASLSGFLN
MAV_5183|M.avium_104 MPLWMQSNKQVSPAGNAAVGLSMSGGSALILAAYYPQQFPYAASLSGFLN
TH_2078|M.thermoresistible__bu LPAYLASNYGVDRNRNAAVGLSMAGSAALVLAIYYPQQFQYAASLSGFLN
Mb1918c|M.bovis_AF2122/97 LPQWLSANRAVKPTGSAAIGLSMAGSSAMILAAYHPQQFIYAGSLSALLD
Rv1886c|M.tuberculosis_H37Rv LPQWLSANRAVKPTGSAAIGLSMAGSSAMILAAYHPQQFIYAGSLSALLD
MLBr_00097|M.leprae_Br4923 LPQYLQSNKQIKPTGSAAVGLSMAGLSALTLAIYHPDQFIYVGSMSGLLD
MAB_1579|M.abscessus_ATCC_1997 LPQWLGVNKGISPAANAVVGLSMAGPSALTLAIYHPQQFVYAGALSAPLH
:* :: * :. .*.:****:* ::: * ::*:** *..::*. *.
Mflv_3335|M.gilvum_PYR-GCK PSEGWWPMLIGLAMNDAGGFNAESMWGPSSDP--AWKRNDPMVNINQLVA
Mvan_3061|M.vanbaalenii_PYR-1 PSEGWWPMLIGLAMNDAGGFNAESMWGPSSDP--AWKRNDPMVNINQLVA
MSMEG_3580|M.smegmatis_MC2_155 PSEGWWPMLIGLAMNDAGGYNAESMWGPSTDP--AWKRNDPMVNINQLVA
MMAR_0328|M.marinum_M PSEGWWPTLIGLAMNDSGGYNANSMWGPSTDP--AWKRNDPMVQIPRLVA
MUL_4793|M.ulcerans_Agy99 PSEGWWPTLIGLAMNDSGGYNANSMWGPSTDP--AWKRNDPMVQIPRLVA
MAV_5183|M.avium_104 PSEGWWPTLIGLAMNDSGGYNANSMWGPSTDP--AWKRNDPMVQIPRLVA
TH_2078|M.thermoresistible__bu LSEGWWPMLVGLAMGDAGGYDPNDMWGRTEDPNSAWKRNDPMVNIDKLVA
Mb1918c|M.bovis_AF2122/97 PSQGMGPSLIGLAMGDAGGYKAADMWGPSSDP--AWERNDPTQQIPKLVA
Rv1886c|M.tuberculosis_H37Rv PSQGMGPSLIGLAMGDAGGYKAADMWGPSSDP--AWERNDPTQQIPKLVA
MLBr_00097|M.leprae_Br4923 PSNAMGPSLIGLAMGDAGGYKAADMWGPSTDP--AWKRNDPTVNVGTLIA
MAB_1579|M.abscessus_ATCC_1997 PSDQKW--QIRVAMSDAGGFNADDMWGPDSDP--AWARNDPFLHIDQLIA
*: : :**.*:**:.. .*** ** ** **** :: *:*
Mflv_3335|M.gilvum_PYR-GCK NNTRIWIYCGTGTPSDLDSGTPGQNLMAAQFLEGFTLRTNVAFRDNYIAA
Mvan_3061|M.vanbaalenii_PYR-1 NNTRIWIYCGTGTPSDLDNGTPGNNLMAAQFLEGFTLRTNIAFRDNYIAA
MSMEG_3580|M.smegmatis_MC2_155 NNTRLWVYCGTGTPSELDAGVSGGNLLAAQFLEGLTLRTNLTFRDQYIAA
MMAR_0328|M.marinum_M NNTRIWVYCGNGTPSDL-----GGDNMPAKFLEGLTLRTNQTFRDTYLAS
MUL_4793|M.ulcerans_Agy99 NNTRIWVYCGNGTPSDL-----GGDNMPAKFLEGLTLRTNQTFRDTYLAS
MAV_5183|M.avium_104 NNTRIWVYCGNGTPSDL-----GGDNVPAKFLEGLTLRTNEQFQNNYAAA
TH_2078|M.thermoresistible__bu NNTRIWVFCGDGTPADIGAGVPEGDNFNAKFLESFTLRTNKTFQQEYIAA
Mb1918c|M.bovis_AF2122/97 NNTRLWVYCGNGTPNEL-----GGANIPAEFLENFVRSSNLKFQDAYNAA
Rv1886c|M.tuberculosis_H37Rv NNTRLWVYCGNGTPNEL-----GGANIPAEFLENFVRSSNLKFQDAYNAA
MLBr_00097|M.leprae_Br4923 NNTRIWMYCGNGKPTEL-----GGNNLPAKLLEGLVRTSNIKFQDGYNAG
MAB_1579|M.abscessus_ATCC_1997 NNTRLWIYCGSGDATDLDKDRNGLEVISGGVIEGQVIDSDKKFAEAYGSA
****:*::** * . :: . . .:*. . :: * : * :.
Mflv_3335|M.gilvum_PYR-GCK GGNNGVFNFPTSGTHSWGYWGQQLQQMKPDIQRVLGA-------------
Mvan_3061|M.vanbaalenii_PYR-1 GGTNGVFNFPTSGTHSWGYWGQQLQQMKPDIQRVLGA-------------
MSMEG_3580|M.smegmatis_MC2_155 GGTNAVFNFPPNGTHTWNYWGQQLLEMKPDIQRVLGA-------------
MMAR_0328|M.marinum_M GGRNGVFNFPTNGTHSWPYWNQQLVAMKGDILKVLNG------PAVPAAP
MUL_4793|M.ulcerans_Agy99 GGRNGVFNFPTNGTHSWPYWNQQLVAMKGDILKVLNG------PAVPAAP
MAV_5183|M.avium_104 GGRNGVFNFPANGTHSWPYWNQQLMAMKPDMQQVLLSGNTTAAPAQPAQP
TH_2078|M.thermoresistible__bu GGKNGVFNFPDAGTHSWGYWGQQLQQMKPDIQRVLGV-------------
Mb1918c|M.bovis_AF2122/97 GGHNAVFNFPPNGTHSWEYWGAQLNAMKGDLQSSLGAG------------
Rv1886c|M.tuberculosis_H37Rv GGHNAVFNFPPNGTHSWEYWGAQLNAMKGDLQSSLGAG------------
MLBr_00097|M.leprae_Br4923 GGHNAVFNFPDSGTHSWEYWGEQLNDMKPDLQQYLGAT------------
MAB_1579|M.abscessus_ATCC_1997 GGANAHFEFPKGGIHNWTYWGNQLRAMKADLVGYLTKPG-----------
** *. *:** * *.* **. ** ** *: *
Mflv_3335|M.gilvum_PYR-GCK ----------QATA
Mvan_3061|M.vanbaalenii_PYR-1 ----------QATV
MSMEG_3580|M.smegmatis_MC2_155 ----------QSAT
MMAR_0328|M.marinum_M AAPEAPAAPVAPAA
MUL_4793|M.ulcerans_Agy99 AAPEAPAAPVAPAA
MAV_5183|M.avium_104 AQPAQPAQPAQPAT
TH_2078|M.thermoresistible__bu -----------SAA
Mb1918c|M.bovis_AF2122/97 --------------
Rv1886c|M.tuberculosis_H37Rv --------------
MLBr_00097|M.leprae_Br4923 -----------PGA
MAB_1579|M.abscessus_ATCC_1997 ----------TAAV