For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGFVEFVKDRWAVLSFLAYQHMSLVLQTLVVAAMCAVFLGVLIYRSPWGVASGNALAAIGLTIPSYALLG VLVGVVGIGVSPSVIMLVFFGSLPILRNVLVGLTGVDQTLVESARGMGMSRLATLVRLELPLAWPVIMTG IRVSAQMLMGTAAIAAFALGPGLGGYIFSGISRMGGANATNSVIAGTVGILILAVILDSVLNVVTRLTTP RGIRV
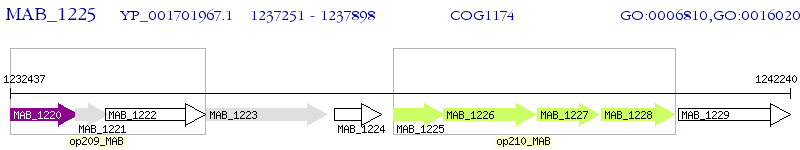
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1225 | - | - | 100% (215) | ABC transporter permease |
| M. abscessus ATCC 19977 | MAB_1227 | - | 2e-30 | 38.76% (209) | ABC transporter permease |
| M. abscessus ATCC 19977 | MAB_0264 | - | 3e-23 | 32.72% (217) | osmoprotectant ABC transporter ProZ |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3783c | proW | 3e-22 | 30.96% (197) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3108 | - | 1e-51 | 46.67% (210) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3757c | proW | 3e-22 | 30.96% (197) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0035 | - | 1e-94 | 80.37% (214) | ABC transporter permease |
| M. avium 104 | MAV_0320 | - | 2e-19 | 29.74% (195) | ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_2927 | - | 2e-98 | 81.40% (215) | ABC transporter, permease protein OpuCB |
| M. thermoresistible (build 8) | TH_0698 | - | 3e-93 | 76.74% (215) | ABC transporter, permease protein OpuCB |
| M. ulcerans Agy99 | MUL_0034 | - | 8e-92 | 78.97% (214) | ABC transporter permease |
| M. vanbaalenii PYR-1 | Mvan_5598 | - | 3e-21 | 33.00% (203) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3783c|M.bovis_AF2122/97 -----------MHYLMTHPG---AAWALTVVHLRLSLLPVLIGLMSAVPL
Rv3757c|M.tuberculosis_H37Rv -----------MHYLMTHPG---AAWALTVVHLRLSLLPVLIGLMSAVPL
MAB_1225|M.abscessus_ATCC_1997 -------MG-FVEFVKDRWA---VLSFLAYQHMSLVLQTLVVAAMCAVFL
MSMEG_2927|M.smegmatis_MC2_155 -------MG-FVEFVQDRWA---VLSFLAYQHMSFVVQTLLIATAAALVV
MMAR_0035|M.marinum_M MEAATEFMG-LYDFVVDRWS---VLWFLAYQHISLVGQTLILATAIALIL
MUL_0034|M.ulcerans_Agy99 -------MG-LYDFVVDRWS---VLWFLAYQHISLVGQALILATAIALIL
TH_0698|M.thermoresistible__bu -------MD-FIEFVRERWS---VLSFLAYQHMSLVVQTLLLSTLLAVAV
Mflv_3108|M.gilvum_PYR-GCK -------MNSLWDYVSTHYQ---QLAFDSYQHVSAVVQSVLLATVIGVAI
MAV_0320|M.avium_104 --MNFVQRAIAYLLSADNWTGPVGLAARILEHLEYTGIAVGASALIAVPV
Mvan_5598|M.vanbaalenii_PYR-1 --MNFLQEALSFIFTAANWGGPAGLSARILEHLQYTVIAVFFSAVIAVPL
. *: .: . .: :
Mb3783c|M.bovis_AF2122/97 GLLVQRAPLLRRLTTATASVIFTIPSLALFVVLPLIIGTRILDEANVIVA
Rv3757c|M.tuberculosis_H37Rv GLLVQRAPLLRRLTTATASVIFTIPSLALFVVLPLIIGTRILDEANVIVA
MAB_1225|M.abscessus_ATCC_1997 GVLIYRSPWGVASGNALAAIGLTIPSYALLGVLVGVVG---IGVSPSVIM
MSMEG_2927|M.smegmatis_MC2_155 GVLIYRSPWASAIGNTLTSVGLTIPSYALLGVLVAFIG---IGVLPSVIM
MMAR_0035|M.marinum_M GVLIYNSPLGVAAGNAVTAVGLTIPSYALLGVLVGIVG---LGVLPSVIM
MUL_0034|M.ulcerans_Agy99 GVLIYNSPLRVAAGNAVTAVGLTIPSYALLGVLVGIVG---LGVLPSVIM
TH_0698|M.thermoresistible__bu GVMLYRSTWGRSLGDSVTSLGLTVPSYALLGVLVGIFG---VGVLPSVVL
Mflv_3108|M.gilvum_PYR-GCK GVLTYRNSLAANLATATSSVILTVPSFALLGLLIPLFG---LGVIPSVAA
MAV_0320|M.avium_104 GLLIGHTGRGTLLVVGAVNGLRALPTLGVLLFGTLLFG---LGLGPSLVA
Mvan_5598|M.vanbaalenii_PYR-1 GMLIGHTGRGTFVVVTGVNALRALPTLGVLLLGVLLWG---LGLIPPTVA
*:: . ::*: .:: . . * :.
Mb3783c|M.bovis_AF2122/97 LAAYTTALLVRAVLEALDAVPAQVHDAATAIGYSRIAQMLKVELPLSIPV
Rv3757c|M.tuberculosis_H37Rv LAAYTTALLVRAVLEALDAVPAQVHDAATAIGYSRIAQMLKVELPLSIPV
MAB_1225|M.abscessus_ATCC_1997 LVFFGSLPILRNVLVGLTGVDQTLVESARGMGMSRLATLVRLELPLAWPV
MSMEG_2927|M.smegmatis_MC2_155 LVFFGFLPILRNVLVGLGGVDRALIESARGMGMSRLSTLLRLEIPLAWPV
MMAR_0035|M.marinum_M LTFFGVFPILRNVVVGLTGVDKGLVESARGMGMSRLTTLVRLELPLAWPV
MUL_0034|M.ulcerans_Agy99 PTFFGVFPILRNVVVGLTGVDKGLVESARGMGMSRLTTLVRLELPLAWPV
TH_0698|M.thermoresistible__bu LTFFGFLPILRNVLVGLNGVDAGLVESARAMGMSRPATLLRLELPLAWPV
Mflv_3108|M.gilvum_PYR-GCK LVLYSLLPIIRNTIVGLNAVDPALTDAARGIGLSRLQTLGRVELRLVWPA
MAV_0320|M.avium_104 LMLLGVPALLAGTYAGIANVEPTVVDAARAMGMTESQVLLRVEVPNALPL
Mvan_5598|M.vanbaalenii_PYR-1 LMLLGIPPLLAGTYSGIANVDRDVVDAARAMGMTESRILLRVETPIAMPL
:: . .: * : ::* .:* :. : ::* *
Mb3783c|M.bovis_AF2122/97 LVAGLRVVAVTNIAMVSVGSVIGIGGLGTWFTAGYQ----TNKSDQIVAG
Rv3757c|M.tuberculosis_H37Rv LVAGLRVVAVTNIAMVSVGSVIGIGGLGTWFTAGYQ----TNKSDQIVAG
MAB_1225|M.abscessus_ATCC_1997 IMTGIRVSAQMLMGTAAIAAFALGPGLGGYIFSGISRMGGANATNSVIAG
MSMEG_2927|M.smegmatis_MC2_155 IMTGVRVSAQMLMGVAAIAAYALGPGLGGYIFSGISRTGGANATNSIVAG
MMAR_0035|M.marinum_M IMTGVRVSAQMLMGIAAIVAFALGPGLGGYIFSGISRMGGANATNSIVAA
MUL_0034|M.ulcerans_Agy99 IMTGVRVSAQMLMGIAAIVAFALGPGLGGYIFSGISRMGGANATNSIVAA
TH_0698|M.thermoresistible__bu IMTGVRISAQMLMGIAAIAAFALGPGLGGYIFSGISRMGGANATNSIIVA
Mflv_3108|M.gilvum_PYR-GCK ILSAMRLSTQMSMGVLAIAAYVKGPGLGNLIFAGLARVGSPTALPMALSG
MAV_0320|M.avium_104 ILGGLRSATLQVVATATVAAYASLGGLGRYLIDGIK----EREFHLALVG
Mvan_5598|M.vanbaalenii_PYR-1 ILGGLRTATLQIVATATVAAYASLGGLGRYLIDGIK----VREFYLALVG
:: .:* : :. :: : *** : * : .
Mb3783c|M.bovis_AF2122/97 VVAMFLLAIVVDVVINLAGRLATPWERAPRAARRRRQVAAPITGGAR---
Rv3757c|M.tuberculosis_H37Rv VVAMFLLAIVVDVVINLAGRLATPWERAPRAARRRRQVAAPITGGAR---
MAB_1225|M.abscessus_ATCC_1997 TVGILILAVILDSVLNVVTRLTTPRGIRV---------------------
MSMEG_2927|M.smegmatis_MC2_155 TVGILILAVVLDTVLNIITRLTTPRGIRV---------------------
MMAR_0035|M.marinum_M TIGILILAVILDTVLNLIARLTTPRGIRA---------------------
MUL_0034|M.ulcerans_Agy99 TIGILILAVILDTVLNVIARLTTPREIRA---------------------
TH_0698|M.thermoresistible__bu TVGILLLAVILDAVLNLVTRLTTPRGIRV---------------------
Mflv_3108|M.gilvum_PYR-GCK TLLIVILALLLDAALVLLGRLTTSKGIR----------------------
MAV_0320|M.avium_104 ALTVAALALVLDGLLAVAVWASVPGTGRLRRTYRPLPHRLAG--------
Mvan_5598|M.vanbaalenii_PYR-1 ALMVTALALILDAALAFAVWASVPGSGRLHRS-GSMPQPLLGDEVALESR
.: : **:::* : . :..
Mb3783c|M.bovis_AF2122/97 ------------------------
Rv3757c|M.tuberculosis_H37Rv ------------------------
MAB_1225|M.abscessus_ATCC_1997 ------------------------
MSMEG_2927|M.smegmatis_MC2_155 ------------------------
MMAR_0035|M.marinum_M ------------------------
MUL_0034|M.ulcerans_Agy99 ------------------------
TH_0698|M.thermoresistible__bu ------------------------
Mflv_3108|M.gilvum_PYR-GCK ------------------------
MAV_0320|M.avium_104 TSYGR-------------------
Mvan_5598|M.vanbaalenii_PYR-1 SRAGRRPDSEARHERGNPSPTVEA