For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTASTITATGAAQSAPATARRPALAARQLLQPSACIVAVMVSLAYVNLADVSASEKRSLGMSNLLVLLGQ HMAISVAATVLTCAVAIPLGIALTRGPVRRYAKTITTLVGFGQAAPAIGLIALGAVLFGIGGIGAVLALT VYGALPIVANTVIGLDGVDTRLVEAARGMGMSARSTLIRVELPLALRVIVAGVRTALVLIVGTAALASFT GAGGLGQLITTGIKLQQPVTLVVGAILVAALALFIDWLARVVEIIAAPRGL
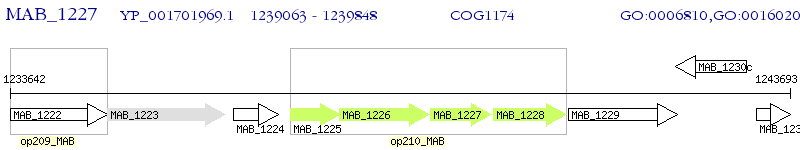
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1227 | - | - | 100% (261) | ABC transporter permease |
| M. abscessus ATCC 19977 | MAB_0264 | - | 1e-33 | 37.69% (199) | osmoprotectant ABC transporter ProZ |
| M. abscessus ATCC 19977 | MAB_1225 | - | 3e-30 | 38.76% (209) | ABC transporter permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3782c | proZ | 4e-28 | 34.17% (199) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3110 | - | 2e-48 | 39.18% (268) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3756c | proZ | 5e-28 | 34.17% (199) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | MLBr_02400 | - | 1e-05 | 25.62% (203) | putative transmembrane protein |
| M. marinum M | MMAR_0033 | - | 6e-99 | 75.40% (248) | hypothetical protein MMAR_0033 |
| M. avium 104 | MAV_0320 | - | 6e-27 | 33.67% (199) | ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_2925 | - | 6e-98 | 78.30% (235) | permease membrane component |
| M. thermoresistible (build 8) | TH_3904 | - | 1e-100 | 74.90% (255) | PUTATIVE conserved hypothetical membrane protein |
| M. ulcerans Agy99 | MUL_0032 | - | 6e-50 | 86.21% (116) | hypothetical protein MUL_0032 |
| M. vanbaalenii PYR-1 | Mvan_5598 | - | 4e-31 | 37.43% (187) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3782c|M.bovis_AF2122/97 -------------------------------------------MNFLQQA
Rv3756c|M.tuberculosis_H37Rv -------------------------------------------MNFLQQA
MAV_0320|M.avium_104 -------------------------------------------MNFVQRA
Mvan_5598|M.vanbaalenii_PYR-1 -------------------------------------------MNFLQEA
MAB_1227|M.abscessus_ATCC_1997 MTASTITATGAAQS---APATARRPALAAR-----QLLQPSACIVAVMVS
MSMEG_2925|M.smegmatis_MC2_155 MTTTAPDTERVALP---AEPSSR--LGLGR-----WLIQPLVCVAAVAGS
TH_3904|M.thermoresistible__bu VTATVDDTGDTGHTGCTGTAGDRASRSAARRWA-RRTVQPVLCLVAVGGA
MMAR_0033|M.marinum_M MSVAGNDINVEGAVKGGAEGPPTAPTTPGRPSVGRWALQPLTCAFAVAGA
MUL_0032|M.ulcerans_Agy99 MGAAATHVCLRGGR-------SAGLCDGGR---------CVRVGAPLAER
Mflv_3110|M.gilvum_PYR-GCK MTATAEVSAEPSTDRESPGTGDPSGTTTAERLR--LLIQPVLVLVVAGAV
MLBr_02400|M.leprae_Br4923 --MRETSNPVFRSLPKQRGGYARFGTDAAPMQQGYYQPDPYAATPYQETR
Mb3782c|M.bovis_AF2122/97 LSYLLTASNWT------GPVGLAVRTCEHLEYTAVAVAASALIAVPVGLL
Rv3756c|M.tuberculosis_H37Rv LSYLLTASNWT------GPVGLAVRTCEHLEYTAVAVAASALIAVPVGLL
MAV_0320|M.avium_104 IAYLLSADNWT------GPVGLAARILEHLEYTGIAVGASALIAVPVGLL
Mvan_5598|M.vanbaalenii_PYR-1 LSFIFTAANWG------GPAGLSARILEHLQYTVIAVFFSAVIAVPLGML
MAB_1227|M.abscessus_ATCC_1997 LAYVNLADVSASEKRSLGMSNLLVLLGQHMAISVAATVLTCAVAIPLGIA
MSMEG_2925|M.smegmatis_MC2_155 LLYVEVADVSESERRALGIGNLLTLLREHMVISVAATVLTCVLAIPLGIA
TH_3904|M.thermoresistible__bu LAYVHLADVSESEKRSLGVPHLLMLLRQHMEISLAATVLTCAIAIPLGVA
MMAR_0033|M.marinum_M LAYVTVADVSESERRSLSVSHLLQLLREHLVISLSATVLTCVVAIPVGIA
MUL_0032|M.ulcerans_Agy99 VASVAAAALTPGDQP---VGHGAHVCGGD---SGGNRIDARTEAALFEAD
Mflv_3110|M.gilvum_PYR-GCK IFWAFNRQLTATQLENINPGNVATLIWQHLLITAAVTAIVLAIGIPLGVL
MLBr_02400|M.leprae_Br4923 AYQVSRPLTIDDVVTRTGMTLAVLVVSAIVSFFLVASNFALATPLTLAGV
.
Mb3782c|M.bovis_AF2122/97 IG--HTGRGTLLVVGAVNGLRALPTLGVLLLGVLLFGLGLGPP-LVALML
Rv3756c|M.tuberculosis_H37Rv IG--HTGRGTLLVVGAVNGLRALPTLGVLLLGVLLFGLGLGPP-LVALML
MAV_0320|M.avium_104 IG--HTGRGTLLVVGAVNGLRALPTLGVLLFGTLLFGLGLGPS-LVALML
Mvan_5598|M.vanbaalenii_PYR-1 IG--HTGRGTFVVVTGVNALRALPTLGVLLLGVLLWGLGLIPP-TVALML
MAB_1227|M.abscessus_ATCC_1997 LTRGPVRRYAKTITTLVGFGQAAPAIGLIALGAVLFGIGGIGA-VLALTV
MSMEG_2925|M.smegmatis_MC2_155 LTRGSMRRYAKPIITVAGFGQAAPAIGLIALGAVLFGIGTTGS-IAALTV
TH_3904|M.thermoresistible__bu LTRGRFRRFATPIITVSGFGQAAPAIGLIALGAVLFGIGAVGA-VAALTV
MMAR_0033|M.marinum_M LTRGSMRRYSKPIITVAGFGQAAPAIGLIALGAVVFGIGQLGA-IVALTV
MUL_0032|M.ulcerans_Agy99 HHRRQFR---------SGRAGYRPDR---AGGGRVRDRSARRD-RCPHRL
Mflv_3110|M.gilvum_PYR-GCK VTRPGAKVLRPLFVGIANVGQAAPAIGLLVLLFLLTAQTGFWIGVLPIAL
MLBr_02400|M.leprae_Br4923 FG-------GLAMVLVATFGGKQNSPAIVLSYAVLEGLFLGAV-SFLFAN
:
Mb3782c|M.bovis_AF2122/97 LGIPSLLASTYAGIASVDPLVVDAARAMGMTESQVLLRVEVPNALPLMLG
Rv3756c|M.tuberculosis_H37Rv LGIPSLLASTYAGIASVDPLVVDAARAMGMTESQVLLRVEVPNALPLMLG
MAV_0320|M.avium_104 LGVPALLAGTYAGIANVEPTVVDAARAMGMTESQVLLRVEVPNALPLILG
Mvan_5598|M.vanbaalenii_PYR-1 LGIPPLLAGTYSGIANVDRDVVDAARAMGMTESRILLRVETPIAMPLILG
MAB_1227|M.abscessus_ATCC_1997 YGALPIVANTVIGLDGVDTRLVEAARGMGMSARSTLIRVELPLALRVIVA
MSMEG_2925|M.smegmatis_MC2_155 YGALPIIANTVIGLDGVDPRLVEAARGMGMSSFSALLKVELPLALRVIVA
TH_3904|M.thermoresistible__bu YGALPIIANTVTGLNGVDDRLIEAARGMGMPTSTTLLRVELPLALPVIIA
MMAR_0033|M.marinum_M YGAMPIIANTVTGLDGVDRRIVEAARGMGLSAFSTLRRVELPLSLPVIVA
MUL_0032|M.ulcerans_Agy99 R-RDPIIANTVTGLDGVDRRIVEAARGMGLSAFSTLRRVELPLSLPVIVA
Mflv_3110|M.gilvum_PYR-GCK YSLLPVLSSTILGIDQVDRSLLDAGLGQGMSRRSLLLRVELPLAVPYILA
MLBr_02400|M.leprae_Br4923 FQVSTVNAGVLIGEAVLG--TFGVFFGMLVVYKTGAIRVTPKFTR-MLVA
.: :.. * : . . . : :* : ::.
Mb3782c|M.bovis_AF2122/97 GLRSATLQVVATATVAAYASLGGLGGYLIDGIKERRFHIALVGAMMVAAL
Rv3756c|M.tuberculosis_H37Rv GLRSATLQVVATATVAAYASLGGLGGYLIDGIKERRFHIALVGAMMVAAL
MAV_0320|M.avium_104 GLRSATLQVVATATVAAYASLGGLGRYLIDGIKEREFHLALVGALTVAAL
Mvan_5598|M.vanbaalenii_PYR-1 GLRTATLQIVATATVAAYASLGGLGRYLIDGIKVREFYLALVGALMVTAL
MAB_1227|M.abscessus_ATCC_1997 GVRTALVLIVGTAALASFTGAGGLGQLITTGIKLQQPVTLVVGAILVAAL
MSMEG_2925|M.smegmatis_MC2_155 GVRTALVLIVGTASLAAFTGGGGLGQLITVGIRLQQTTTLVVGAILVASL
TH_3904|M.thermoresistible__bu GVRTALVLIVGTAALASFTGAGGLGQLITTGIKLQQTVTLIVGAVLVAAL
MMAR_0033|M.marinum_M GVRTALVLIVGTAALASFTGGGGLGQLITTGIKLQQPVTLIVGAILVAAL
MUL_0032|M.ulcerans_Agy99 GVRTALVLIVGTAALASFTGGRGLGQLITTGIKLQQPVTLIVGAILVAAL
Mflv_3110|M.gilvum_PYR-GCK GLRTSLVLAVGTATLSFLVNAGGLGILIDTGYKLQDNVTLILGSVLAVCL
MLBr_02400|M.leprae_Br4923 GTFGVLALMLGNFVLAMFGLGGGTGLGLRSGGPVAILFSLACIGIAAFGF
* :.. :: * * : * .: . :
Mb3782c|M.bovis_AF2122/97 ALTLDGLLALAGWVSVPGTGRMRKLAAVVDKPAAGGGHALR---------
Rv3756c|M.tuberculosis_H37Rv ALTLDGLLALAGWVSVPGTGRMRKLAAVVDKPAAGGGHALR---------
MAV_0320|M.avium_104 ALVLDGLLAVAVWASVPGTGRLRRTYRPLPHRLAGTSYGR----------
Mvan_5598|M.vanbaalenii_PYR-1 ALILDAALAFAVWASVPGSGRLHR-SGSMPQPLLGDEVALESRSRAGRRP
MAB_1227|M.abscessus_ATCC_1997 ALFIDWLARVVEIIAAPRGL------------------------------
MSMEG_2925|M.smegmatis_MC2_155 ALFIDWLARVVEMLAAPKGLG-----------------------------
TH_3904|M.thermoresistible__bu ALFIDWLARMVEVVAAPKGL------------------------------
MMAR_0033|M.marinum_M ALFIDWLARLIEMVAAPRGL------------------------------
MUL_0032|M.ulcerans_Agy99 ALFIDWLARLIEMVAAPRGL------------------------------
Mflv_3110|M.gilvum_PYR-GCK ALLVDWLGGLAEFFLSPKGLR-----------------------------
MLBr_02400|M.leprae_Br4923 LLDFDAADQMIRAGAPEKAAWGIALGLTVTLVWLYVEILRLLSYLQND--
* .* .
Mb3782c|M.bovis_AF2122/97 -----------------
Rv3756c|M.tuberculosis_H37Rv -----------------
MAV_0320|M.avium_104 -----------------
Mvan_5598|M.vanbaalenii_PYR-1 DSEARHERGNPSPTVEA
MAB_1227|M.abscessus_ATCC_1997 -----------------
MSMEG_2925|M.smegmatis_MC2_155 -----------------
TH_3904|M.thermoresistible__bu -----------------
MMAR_0033|M.marinum_M -----------------
MUL_0032|M.ulcerans_Agy99 -----------------
Mflv_3110|M.gilvum_PYR-GCK -----------------
MLBr_02400|M.leprae_Br4923 -----------------