For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNTGNTDVWLVIPAYNEGPVIADVIAAALGTFPNIVCVDDGSVDRTAALAHGAGAHVVRHPVNLGQGAAI QTGVEYARAQPGAQIFVTFDADGQHQVKDVVAMVDRLRLEPLDIIVGTRFGAGSGPGHVPLIKRIVLKTV VLLSPRTRRLNLTDAHNGLRVFNRTVANGLNITMNGMGHASEFIAMISENHWRVAEQPVDILYTEYSMAK GQSLINGVNIMFDTAFRRRLP*
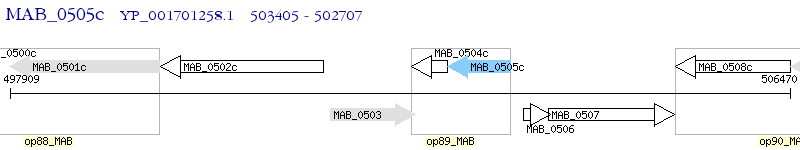
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0505c | - | - | 100% (232) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_2207 | - | 9e-08 | 25.11% (219) | polyprenol phosphate mannosyl transferase 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3655 | - | 1e-83 | 62.82% (234) | transferase |
| M. gilvum PYR-GCK | Mflv_3118 | - | 3e-09 | 22.64% (212) | dolichyl-phosphate beta-D-mannosyltransferase |
| M. tuberculosis H37Rv | Rv3631 | - | 1e-83 | 62.82% (234) | transferase |
| M. leprae Br4923 | MLBr_00207 | - | 2e-79 | 60.43% (230) | putative glycosyltransferase |
| M. marinum M | MMAR_5131 | - | 4e-85 | 65.37% (231) | glycosyl transferase |
| M. avium 104 | MAV_0525 | - | 4e-82 | 64.25% (221) | glycosyl transferase |
| M. smegmatis MC2 155 | MSMEG_3859 | - | 7e-11 | 26.13% (222) | glycosyl transferase, group 2 family protein |
| M. thermoresistible (build 8) | TH_0733 | - | 2e-08 | 26.02% (196) | Polyprenol-monophosphomannose synthase Ppm1 |
| M. ulcerans Agy99 | MUL_4208 | - | 3e-85 | 65.37% (231) | glycosyl transferase |
| M. vanbaalenii PYR-1 | Mvan_0217 | - | 8e-09 | 27.46% (193) | glycosyl transferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3859|M.smegmatis_MC2_155 -----------------MSVPGEREQGAGEDPATVRPTQRTLVIIPTYNE
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK -----------------MSVPG---GVTGREEPAGRPSARTLVIIPTYNE
MLBr_00207|M.leprae_Br4923 -------------------------------MGIETCYPGVWIVIPAFNE
MAV_0525|M.avium_104 -------------------------------------------MIPAFNE
Mb3655|M.bovis_AF2122/97 ---------------------------MASKMDTETHYSDVWVVIPAFNE
Rv3631|M.tuberculosis_H37Rv ---------------------------MASKMDTETHYSDVWVVIPAFNE
MMAR_5131|M.marinum_M -------------------------------MDIDTHHPDVWIVVPAFNE
MUL_4208|M.ulcerans_Agy99 ---------------------------MAVKMDIDTHHPDVWIVVPAFNE
MAB_0505c|M.abscessus_ATCC_199 --------------------------------MSNTGNTDVWLVIPAYNE
Mvan_0217|M.vanbaalenii_PYR-1 MGELPATGHVGSASHQGQTTTSSPHAQFMLARDGVKVTPSVSLVIPVRNE
MSMEG_3859|M.smegmatis_MC2_155 RENLPLIVGRVHHACPQVHILVVDDGSPDGTGALADELALADPDRVHVMH
TH_0733|M.thermoresistible__bu -----------------VHVLIVDDGSPDGTGELADELALADPDRVHVMH
Mflv_3118|M.gilvum_PYR-GCK RDNLLSIVSRVNDAAPDVHVLIVDDGSPDGTGELADELALGDPDRIHVMH
MLBr_00207|M.leprae_Br4923 AAIIGEVVTDVRAVFD--HVVCVDDGSADGTGEIALRAGA------HLVR
MAV_0525|M.avium_104 AAVIGEVIADVRSVFD--NVVCVDDGSTDDTGEIARRAGA------HLVR
Mb3655|M.bovis_AF2122/97 AAVIGKVVTDVRSVFD--HVVCVDDGSTDGTGDIARRSGA------HLVR
Rv3631|M.tuberculosis_H37Rv AAVIGKVVTDVRSVFD--HVVCVDDGSTDGTGDIARRSGA------HLVR
MMAR_5131|M.marinum_M ATVIGEVIAQLRSTFG--HVVCVDDGSTDGTGDIALRAGA------HVVA
MUL_4208|M.ulcerans_Agy99 ATVIGEVIAQLRSTFG--HVVCVDDGSTDGTGDIALRAGA------HVVA
MAB_0505c|M.abscessus_ATCC_199 GPVIADVIAAALGTFP--NIVCVDDGSVDRTAALAHGAGA------HVVR
Mvan_0217|M.vanbaalenii_PYR-1 ARNVAWVLEQITEDVD--EIILVDGASTDATLITALRYRP-----DIKVV
.:: **..* * * * :
MSMEG_3859|M.smegmatis_MC2_155 RTSKAGLGAAYLAGFDWGLRR-GYSVLVEMDADGSHAPEELSRLLDAVDA
TH_0733|M.thermoresistible__bu RASKAGLGAAYLAGFRWGLNR-QYSVLVEMDADGSHAPEELHRLLDAVDA
Mflv_3118|M.gilvum_PYR-GCK RTSKDGLGAAYLAGFAWGLNR-EYTVLVEMDADGSHAPEDLHRLLAEIDA
MLBr_00207|M.leprae_Br4923 HPVNLGQGAAIQTGVEYARRQPAAQVFATFDADGQHRVKDLAALVDRLGA
MAV_0525|M.avium_104 HPINLGQGAAIQTGVEYARRQPGAQVFATFDADGQHRVKDLAAMVNRLRV
Mb3655|M.bovis_AF2122/97 HPINLGQGAAIQTGIEYARKQPGAQVFATFDGDGQHRVKDVAAMVDRLGA
Rv3631|M.tuberculosis_H37Rv HPINLGQGAAIQTGIEYARKQPGAQVFATFDGDGQHRVKDVAAMVDRLGA
MMAR_5131|M.marinum_M HPINLGQGAAIQTGVEYARKQPGAQIFATFDADGQHRVKDLAAMVHRLSV
MUL_4208|M.ulcerans_Agy99 HPINLGQGAAIQTGVEYARKQPGAQIFATFDADGQHRVKDLAAMVHRLSV
MAB_0505c|M.abscessus_ATCC_199 HPVNLGQGAAIQTGVEYARAQPGAQIFVTFDADGQHQVKDVVAMVDRLRL
Mvan_0217|M.vanbaalenii_PYR-1 AQDGVGKGSALRTGFFAASGD----IIVMMDADGSMAPQEIRHYVHFLRN
* *:* :*. . ::. :*.**. ::: : :
MSMEG_3859|M.smegmatis_MC2_155 G-ADLAIGSRYVPGGTVR-----NWPWRRLVLSKTANTYSRFLLGVGIHD
TH_0733|M.thermoresistible__bu G-ADLAIGSRYVDGGQVR-----NWPRRRLLLSRTANTYSRLLLGVDIRD
Mflv_3118|M.gilvum_PYR-GCK G-ADVVIGSRYVEGGDVR-----NWPRRRMVLSRTANAYSRILLGVDIHD
MLBr_00207|M.leprae_Br4923 HDVDVVIGTRFGRLDGGRLPILKGPPFLKRVVLRTAARLSRRGRRLGLTD
MAV_0525|M.avium_104 GDVDVVIGTRFGTRQGAR------PPLVKRMVLQTAARLSRRGRRLGLTD
Mb3655|M.bovis_AF2122/97 GDVDVVIGTRFGRPVGKASA--SRPPLMKRIVLQTGARLSRRGRRLGLTD
Rv3631|M.tuberculosis_H37Rv GDVDVVIGTRFGRPVGKASA--SRPPLMKRIVLQTGARLSRRGRRLGLTD
MMAR_5131|M.marinum_M GDVDVVIGTRFGRPVG------SRPPLLKRIVLQTAARLSPRGRRLGLTD
MUL_4208|M.ulcerans_Agy99 GDVDVVIGTRFGRPVG------SRPPLLKRIVLQTAARLSPRGRRLGLTD
MAB_0505c|M.abscessus_ATCC_199 EPLDIIVGTRFGAGSGPG-----HVPLIKRIVLKTVVLLSPRTRRLNLTD
Mvan_0217|M.vanbaalenii_PYR-1 G-YDFVKGSRFIGGGGSL-----DITAFRKLGNRFLLGVFNSLYDSDLTD
*. *:*: . : : : .: *
MSMEG_3859|M.smegmatis_MC2_155 ITAGYRAYRREVLEKIDLSAVDSKGYCFQIDLTWRAINNGFSVVEVPITF
TH_0733|M.thermoresistible__bu ITAGYRAYRREVLEKIDLDAVDSKGYCFQIDLTWRAINHGFKVVEVPITF
Mflv_3118|M.gilvum_PYR-GCK ITAGYRAYRREVLEKIDLAAVDSKGYGFQVDLTWRSINAGFTVVEVPITF
MLBr_00207|M.leprae_Br4923 TNNGLRVFNKKVADGLDITMT---GMSHANEFVMLIADNHWRVDEVPVEV
MAV_0525|M.avium_104 TNNGLRVFNKKVADGLDITMS---GMSHANEFVMLIDENHWRVVEEPIEV
Mb3655|M.bovis_AF2122/97 TNNGLRVFNKTVADGLNITMS---GMSHATEFIMLIAENHWRVAEEPVEV
Rv3631|M.tuberculosis_H37Rv TNNGLRVFNKTVADGLNITMS---GMSHATEFIMLIAENHWRVAEEPVEV
MMAR_5131|M.marinum_M TNNGLRVFNKTVADGLNITMS---GMSHATEFVMLIAENHWRVAEEPVEV
MUL_4208|M.ulcerans_Agy99 TNNGLRVFNKTVADGLNITMS---GMSHATEFVMLIAENHWRVAEEPVEV
MAB_0505c|M.abscessus_ATCC_199 AHNGLRVFNRTVANGLNITMN---GMGHASEFIAMISENHWRVAEQPVDI
Mvan_0217|M.vanbaalenii_PYR-1 LCYGFCAFHRRYLELLELSAT---GFEIEAEMTVRAMQAGLRIAEVPSLE
* .:.: : ::: * :: : : * *
MSMEG_3859|M.smegmatis_MC2_155 TERELGVSKM----SGSNIREAMFKVAEWGIRGRLDRARGVVR-------
TH_0733|M.thermoresistible__bu TERELGQSKM----SGSNIREAIFKVAEWGIRGRLDRARGIVR-------
Mflv_3118|M.gilvum_PYR-GCK TEREHGVSKM----DGSTIREAIIKVAQWGMRARIDRARGNTP-------
MLBr_00207|M.leprae_Br4923 LYTEYSKS------KGQPLLNGVNIIFDGFLRGRMPK-------------
MAV_0525|M.avium_104 LYTEYSRS------KGQPLLNGVNIIFDGFLRGRIPR-------------
Mb3655|M.bovis_AF2122/97 LYTEYSKS------KGQPLLNGVNIIFDGFLRGRMPR-------------
Rv3631|M.tuberculosis_H37Rv LYTEYSKS------KGQPLLNGVNIIFDGFLRGRMPR-------------
MMAR_5131|M.marinum_M LYTDYSKS------KGQPLLNGVNIIFDGFLRGRIRR-------------
MUL_4208|M.ulcerans_Agy99 LYTDYSKS------KGQPLLNGVNIIFDGFLRGRIRR-------------
MAB_0505c|M.abscessus_ATCC_199 LYTEYSMA------KGQSLINGVNIMFDTAFRRRLP--------------
Mvan_0217|M.vanbaalenii_PYR-1 MPRRSGKSNLRAVRDGTRVLRTVLHGHRSGVSGHLLEVLSQRRGPAAAAA
. : .* : . : . ::
MSMEG_3859|M.smegmatis_MC2_155 ---
TH_0733|M.thermoresistible__bu ---
Mflv_3118|M.gilvum_PYR-GCK ---
MLBr_00207|M.leprae_Br4923 ---
MAV_0525|M.avium_104 ---
Mb3655|M.bovis_AF2122/97 ---
Rv3631|M.tuberculosis_H37Rv ---
MMAR_5131|M.marinum_M ---
MUL_4208|M.ulcerans_Agy99 ---
MAB_0505c|M.abscessus_ATCC_199 ---
Mvan_0217|M.vanbaalenii_PYR-1 EAC