For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SVPGEREQGAGEDPATVRPTQRTLVIIPTYNERENLPLIVGRVHHACPQVHILVVDDGSPDGTGALADEL ALADPDRVHVMHRTSKAGLGAAYLAGFDWGLRRGYSVLVEMDADGSHAPEELSRLLDAVDAGADLAIGSR YVPGGTVRNWPWRRLVLSKTANTYSRFLLGVGIHDITAGYRAYRREVLEKIDLSAVDSKGYCFQIDLTWR AINNGFSVVEVPITFTERELGVSKMSGSNIREAMFKVAEWGIRGRLDRARGVVR*
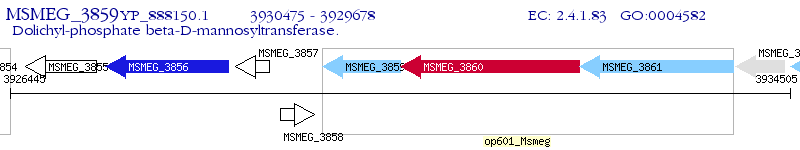
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3859 | - | - | 100% (265) | glycosyl transferase, group 2 family protein |
| M. smegmatis MC2 155 | MSMEG_0995 | - | 1e-18 | 33.33% (225) | glycosyl transferase, family protein 2 |
| M. smegmatis MC2 155 | MSMEG_4987 | - | 3e-09 | 28.71% (209) | glycosyl transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2077c | ppm1 | 1e-115 | 82.64% (242) | polyprenol-monophosphomannose synthase Ppm1 |
| M. gilvum PYR-GCK | Mflv_3118 | - | 1e-113 | 76.60% (265) | dolichyl-phosphate beta-D-mannosyltransferase |
| M. tuberculosis H37Rv | Rv2051c | ppm1 | 1e-115 | 82.64% (242) | polyprenol-monophosphomannose synthase Ppm1 |
| M. leprae Br4923 | MLBr_01440 | - | 1e-108 | 75.72% (243) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_2207 | - | 1e-114 | 78.97% (252) | polyprenol phosphate mannosyl transferase 1 |
| M. marinum M | MMAR_3028 | ppm1B | 1e-118 | 78.74% (254) | polyprenol-monophosphomannose synthase, Ppm1B |
| M. avium 104 | MAV_4605 | - | 4e-18 | 34.10% (217) | glycosyltransferase |
| M. thermoresistible (build 8) | TH_0733 | - | 1e-109 | 89.77% (215) | Polyprenol-monophosphomannose synthase Ppm1 |
| M. ulcerans Agy99 | MUL_2269 | ppm1B | 1e-118 | 78.35% (254) | C-term polyprenol-monophosphomannose synthase Ppm1B |
| M. vanbaalenii PYR-1 | Mvan_3418 | - | 1e-115 | 77.74% (265) | dolichyl-phosphate beta-D-mannosyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 MKLGAWVAAQLPTTRTAVRTRLTRLVVSIVAGLLLYASFPPRNCWWAAVV
Rv2051c|M.tuberculosis_H37Rv MKLGAWVAAQLPTTRTAVRTRLTRLVVSIVAGLLLYASFPPRNCWWAAVV
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 ALALLAWVLTHRATTPVGGLGYGLLFGLVFYVSLLPWIGELVGPGPWLAL
Rv2051c|M.tuberculosis_H37Rv ALALLAWVLTHRATTPVGGLGYGLLFGLVFYVSLLPWIGELVGPGPWLAL
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 ATTCALFPGIFGLFAVVVRLLPGWPIWFAVGWAAQEWLKSILPFGGFPWG
Rv2051c|M.tuberculosis_H37Rv ATTCALFPGIFGLFAVVVRLLPGWPIWFAVGWAAQEWLKSILPFGGFPWG
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 SVAFGQAEGPLLPLVQLGGVALLSTGVALVGCGLTAIALEIEKWWRTGGQ
Rv2051c|M.tuberculosis_H37Rv SVAFGQAEGPLLPLVQLGGVALLSTGVALVGCGLTAIALEIEKWWRTGGQ
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 GDAPPAVVLPAACICLVLFAAIVVWPQVRHAGSGSGGEPTVTVAVVQGNV
Rv2051c|M.tuberculosis_H37Rv GDAPPAVVLPAACICLVLFAAIVVWPQVRHAGSGSGGEPTVTVAVVQGNV
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 PRLGLDFNAQRRAVLDNHVEETLRLAADVHAGLAQQPQFVIWPENSSDID
Rv2051c|M.tuberculosis_H37Rv PRLGLDFNAQRRAVLDNHVEETLRLAADVHAGLAQQPQFVIWPENSSDID
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 PFVNPDAGQRISAAAEAIGAPILIGTLMDVPGRPRENPEWTNTAIVWNPG
Rv2051c|M.tuberculosis_H37Rv PFVNPDAGQRISAAAEAIGAPILIGTLMDVPGRPRENPEWTNTAIVWNPG
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 TGPADRHDKAIVQPFGEYLPMPWLFRHLSGYADRAGHFVPGNGTGVVRIA
Rv2051c|M.tuberculosis_H37Rv TGPADRHDKAIVQPFGEYLPMPWLFRHLSGYADRAGHFVPGNGTGVVRIA
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 GVPVGVATCWEVIFDRAPRKSILGGAQLLTVPSNNATFNKTMSEQQLAFA
Rv2051c|M.tuberculosis_H37Rv GVPVGVATCWEVIFDRAPRKSILGGAQLLTVPSNNATFNKTMSEQQLAFA
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 KVRAVEHDRYVVVAGTTGISAVIAPDGGELIRTDFFQPAYLDSQVRLKTR
Rv2051c|M.tuberculosis_H37Rv KVRAVEHDRYVVVAGTTGISAVIAPDGGELIRTDFFQPAYLDSQVRLKTR
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------------------
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M --------------------------------------------------
MUL_2269|M.ulcerans_Agy99 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 LTPATRWGPILQWILVGAAAAVVLVAMRQNGWFPRPRRSEPKGENDDSDA
Rv2051c|M.tuberculosis_H37Rv LTPATRWGPILQWILVGAAAAVVLVAMRQNGWFPRPRRSEPKGENDDSDA
MLBr_01440|M.leprae_Br4923 --------------------------------------------------
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 --------------------------------------MSVPGEREQGAG
TH_0733|M.thermoresistible__bu --------------------------------------------------
Mflv_3118|M.gilvum_PYR-GCK --------------------------------------MSVPGG--VTGR
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------MSVPGG--MTDG
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3028|M.marinum_M -------------------------------------------MTTGQPA
MUL_2269|M.ulcerans_Agy99 -------------------------------------------MTTGQPA
Mb2077c|M.bovis_AF2122/97 PPGRSEASGPPALSESDDELIQPEQGGRHSSGFGRHRATSRSYMTTGQPA
Rv2051c|M.tuberculosis_H37Rv PPGRSEASGPPALSESDDELIQPEQGGRHSSGFGRHRATSRSYMTTGQPA
MLBr_01440|M.leprae_Br4923 -------------------------------------------MITGQPA
MAV_4605|M.avium_104 --------------------------------------------------
MSMEG_3859|M.smegmatis_MC2_155 EDPATVRPTQRTLVIIPTYNERENLPLIVGRVHHACPQVHILVVDDGSPD
TH_0733|M.thermoresistible__bu --------------------------------------VHVLIVDDGSPD
Mflv_3118|M.gilvum_PYR-GCK EEPAG-RPSARTLVIIPTYNERDNLLSIVSRVNDAAPDVHVLIVDDGSPD
Mvan_3418|M.vanbaalenii_PYR-1 PGPGT-RPSDRTLVIIPTYNERQNLPLIVGRVHQARPDVDILIVDDDSPD
MAB_2207|M.abscessus_ATCC_1997 MDPVTERPSARTLVIIPTYNERENLPLILGRLHKAQPDVHVLIVDDGSPD
MMAR_3028|M.marinum_M PQSPGPRPSERTLVIIPTYNERENLPLIHRRVVDACPHVHVLIVDDNSPD
MUL_2269|M.ulcerans_Agy99 PQSPGPRPSERTLVIIPTYNERENLPLIHRRVVDACPHVHVLIVDDNSPD
Mb2077c|M.bovis_AF2122/97 PPAPGNRPSQRVLVIIPTFNERENLPVIHRRLTQACPAVHVLVVDDSSPD
Rv2051c|M.tuberculosis_H37Rv PPAPGNRPSQRVLVIIPTFNERENLPVIHRRLTQACPAVHVLVVDDSSPD
MLBr_01440|M.leprae_Br4923 RKISGNRPSQCVLVIIPTFNELENLPVTHGRLKDAYPEIHVLIVDDGSPD
MAV_4605|M.avium_104 ----MPGADGRVTVVLPCLNEEESLPGVLATIP---PGYRALVVDNNSTD
*:**:.*.*
MSMEG_3859|M.smegmatis_MC2_155 GTGALADELALADPDRVHVMHRTSKAGLGAAYLAGFDWGLRRGYSVLVEM
TH_0733|M.thermoresistible__bu GTGELADELALADPDRVHVMHRASKAGLGAAYLAGFRWGLNRQYSVLVEM
Mflv_3118|M.gilvum_PYR-GCK GTGELADELALGDPDRIHVMHRTSKDGLGAAYLAGFAWGLNREYTVLVEM
Mvan_3418|M.vanbaalenii_PYR-1 GTGQLADELSVADPDRIHVMHRTAKEGLGAAYLAGFAWGLGRQYTVLVEM
MAB_2207|M.abscessus_ATCC_1997 GTGELADEAALADPDRVHVMHRKEKGGLGAAYIAGFGWGLARQYSVLVEM
MMAR_3028|M.marinum_M GTGRLADELVAADPDRIHVLHRTAKNGLGAAYLEGFAWGLSRGYSVLVEM
MUL_2269|M.ulcerans_Agy99 GTGRLADELVAADPDRIHVLHRTAKNGLGAAYLEGFAWGLSRGYSVLVEM
Mb2077c|M.bovis_AF2122/97 GTGQLADELAQADPGRTHVMHRTAKNGLGAAYLAGFAWGLSREYSVLVEM
Rv2051c|M.tuberculosis_H37Rv GTGQLADELAQADPGRTHVMHRTAKNGLGAAYLAGFAWGLSREYSVLVEM
MLBr_01440|M.leprae_Br4923 GTGELADELAQVDPGCTHVMHRTTKDGLGTAYLAGFAWGMSRDYSVLVEM
MAV_4605|M.avium_104 DTAGVARRHGAQ-------VVTETRAGYGAAVHAG---VLAATTPIVAVL
.*. :* . : : * *:* * : .::. :
MSMEG_3859|M.smegmatis_MC2_155 DADGSHAPEELSRLLDAVDAGADLAIGSRYVPGGTVRNWPWRRLVLSKTA
TH_0733|M.thermoresistible__bu DADGSHAPEELHRLLDAVDAGADLAIGSRYVDGGQVRNWPRRRLLLSRTA
Mflv_3118|M.gilvum_PYR-GCK DADGSHAPEDLHRLLAEIDAGADVVIGSRYVEGGDVRNWPRRRMVLSRTA
Mvan_3418|M.vanbaalenii_PYR-1 DADGSHPPEQLARLLDAVDAGADVVIGSRYVSGGEVVNWPRRRLVLSRTA
MAB_2207|M.abscessus_ATCC_1997 DADGSHAPEQLSRLLDAVDAGADLAIGSRYVPGGTVVNWPWRRLVLSRSA
MMAR_3028|M.marinum_M DADGSHAPEQLHRLLDAVDAGADLSIGSRYVPGGTVRNWPWRRLALSKTA
MUL_2269|M.ulcerans_Agy99 DADGSHAPEQLHRLLDAVDAGADLSIGSRYVPGGTVRNWPWQRLALSKTA
Mb2077c|M.bovis_AF2122/97 DADGSHAPEQLQRLLDAVDAGADLAIGSRYVAGGTVRNWPWRRLVLSKTA
Rv2051c|M.tuberculosis_H37Rv DADGSHAPEQLQRLLDAVDAGADLAIGSRYVAGGTVRNWPWRRLVLSKTA
MLBr_01440|M.leprae_Br4923 DADGSHAPEQLHRLLGAVDAGADLAIGSRYVNGGTVRNWPWQRLALSKTA
MAV_4605|M.avium_104 DADGSMDGADLPRLVAALEAGADLVTGRRRPVRG--LRWPWVARVGTVVM
***** :* **: ::****: * * * .** :
MSMEG_3859|M.smegmatis_MC2_155 NTYSRFLLGVGIHDITAGYRAYRREVLEKIDLSAVDSKGYCFQIDLTWRA
TH_0733|M.thermoresistible__bu NTYSRLLLGVDIRDITAGYRAYRREVLEKIDLDAVDSKGYCFQIDLTWRA
Mflv_3118|M.gilvum_PYR-GCK NAYSRILLGVDIHDITAGYRAYRREVLEKIDLAAVDSKGYGFQVDLTWRS
Mvan_3418|M.vanbaalenii_PYR-1 NGYSRILLGVDIHDITAGYRAYRREVLEKIDLAAVDTKGYGFQVDLTWRS
MAB_2207|M.abscessus_ATCC_1997 NVYARLVLGVKPHDITAGYRAYRREVLEKLDLGAVESHGYCFQIDLTLRT
MMAR_3028|M.marinum_M NTYSRLALGIGVHDITAGYRAYRREVLEKIDLASVDSKGYCFQIEMTWRS
MUL_2269|M.ulcerans_Agy99 NTYSRLALGIGVHDITAGYRAYRREVLEKIDLASVDSKGYCFQIEMTWRS
Mb2077c|M.bovis_AF2122/97 NTYSRLALGIGIHDITAGYRAYRREALEAIDLDGVDSKGYCFQIDLTWRT
Rv2051c|M.tuberculosis_H37Rv NTYSRLALGIGIHDITAGYRAYRREALEAIDLDGVDSKGYCFQIDLTWRT
MLBr_01440|M.leprae_Br4923 NKYSRLALGIDVHDITAGYRAYRREVLEAIDLDSVASKGYCFQIDLTWRT
MAV_4605|M.avium_104 SWRLRTRHGLPVHDIAP-MRVARRDALLGLGVADRRS---GYPLELLVRA
. * *: :**:. *. **:.* :.: : : ::: *:
MSMEG_3859|M.smegmatis_MC2_155 INNGFSVVEVPITFTERELGVSKMSGSNIREAMFKVAEWGIRGRLDRARG
TH_0733|M.thermoresistible__bu INHGFKVVEVPITFTERELGQSKMSGSNIREAIFKVAEWGIRGRLDRARG
Mflv_3118|M.gilvum_PYR-GCK INAGFTVVEVPITFTEREHGVSKMDGSTIREAIIKVAQWGMRARIDRARG
Mvan_3418|M.vanbaalenii_PYR-1 INAGFTVIEVPITFTEREHGVSKMDGSTIREAIIKVAQWGLRARLDRARG
MAB_2207|M.abscessus_ATCC_1997 IAHGFEVAEVPITFTERAIGESKMSGSIINEALVKVTKWGVQSRLDRARG
MMAR_3028|M.marinum_M VNSGFVVVEVPITFTEREIGVSKMSGSNIREALFKVTRWGVKGRFAGSEG
MUL_2269|M.ulcerans_Agy99 VNSGFVVVEVPITFTEREIGVSKMSGSNIREALFKVTRWGVKGRFAGSEG
Mb2077c|M.bovis_AF2122/97 VSNGFVVTEVPITFTERELGVSKMSGSNIREALVKVARWGIEGRLSRSDH
Rv2051c|M.tuberculosis_H37Rv VSNGFVVTEVPITFTERELGVSKMSGSNIREALVKVARWGIEGRLSRSDH
MLBr_01440|M.leprae_Br4923 VNNGFVIIEVPITFTEREFGLSKMSGSNIREALVKVTRWGIDGRIQRART
MAV_4605|M.avium_104 AAAGWRVVELDVRYGPRTGGKSKVSGSLRGSITAILDFWKVIS-------
*: : *: : : * * **:.** . : * : .
MSMEG_3859|M.smegmatis_MC2_155 VVR---------------------
TH_0733|M.thermoresistible__bu IVR---------------------
Mflv_3118|M.gilvum_PYR-GCK NTP---------------------
Mvan_3418|M.vanbaalenii_PYR-1 TTR---------------------
MAB_2207|M.abscessus_ATCC_1997 VNR---------------------
MMAR_3028|M.marinum_M GGDRAHTHDHS-------------
MUL_2269|M.ulcerans_Agy99 GGDRAHTHDHS-------------
Mb2077c|M.bovis_AF2122/97 ARARPDIARPGAGGSRVSRADVTE
Rv2051c|M.tuberculosis_H37Rv ARARPDIARPGAGGSRVSRADVTE
MLBr_01440|M.leprae_Br4923 GRAYNSVSRGGGTGSSPSAP----
MAV_4605|M.avium_104 ------------------------