For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MITTKTVNGVQIAFDDQGHEPGPVFVTLSGWAHDLRAYDGMLPYLRAAQRTVRVCWRGHGPDRNLVGDFG IDEMAADTIGLLDALEVDSFVPIAHAHGGWAALEIADRLGAQRVPAVMILDLIMTPAPREFVAALHGIQD PERWKEGRDGLVQSWLAGTTNQAVLDHVRYDSGGHGFDMWARAGRVIDEAYRTWGSPMRRMEALAEPCAI RHVFSHPKIGEYDALHDDFAARHPWFSYRRLGGETHFPGIELPQQVAAEAIDLLAGARI
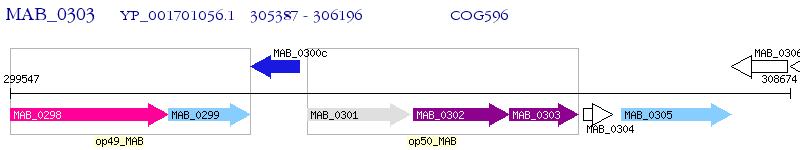
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0303 | - | - | 100% (269) | putative dioxygenase (1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase) |
| M. abscessus ATCC 19977 | MAB_4679 | - | 1e-87 | 56.44% (264) | putative dioxygenase |
| M. abscessus ATCC 19977 | MAB_0679c | - | 7e-09 | 29.20% (137) | hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1348 | - | 2e-07 | 27.42% (124) | 3-oxoadipate enol-lactonase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00862 | ephD | 9e-05 | 29.91% (107) | short chain dehydrogenase |
| M. marinum M | MMAR_3948 | - | 1e-05 | 31.30% (115) | hydrolase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3336 | - | 9e-07 | 28.57% (112) | hydrolase |
| M. thermoresistible (build 8) | TH_3727 | - | 5e-06 | 28.10% (121) | PUTATIVE Chloride peroxidase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1348|M.gilvum_PYR-GCK -----------------MSAVDVHVIESGLPD--GPAVVLSNSLGATHRM
TH_3727|M.thermoresistible__bu -------------MATITTSDGVEIFYRDWGS--GQPIVFSHGWPLTADD
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPE--GPAIVLVHGFPDSHVL
MMAR_3948|M.marinum_M --------------MPIATINGQKIHYTDAGA--GPPVLATHATLMDLIS
MAB_0303|M.abscessus_ATCC_1997 -------------MITTKTVNGVQIAFDDQGHEPGPVFVTLSGWAHDLRA
MSMEG_3336|M.smegmatis_MC2_155 -------------MVERPDETTIRYSITGPSE--GSTLVLIHGWGCERRY
. * .: .
Mflv_1348|M.gilvum_PYR-GCK WDAQLDALERR-FRVLRYDTRGHG--ASPVPDGPYSIDDLADDVVGILDR
TH_3727|M.thermoresistible__bu WDPQMLFFLNAGYRVIAHDRRGHGR-SSPVPDG-HDMDHYADDLAALVEH
MLBr_00862|M.leprae_Br4923 WDGVVPLLAKR-FRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDE
MMAR_3948|M.marinum_M LEPVTTKLVDAGYRVIAFDLRGHG--ETVYDQQPYTYVDVAGDVIGLADY
MAB_0303|M.abscessus_ATCC_1997 YDGMLPYLRAA-QRTVRVCWRGHG--PDRNLVGDFGIDEMAADTIGLLDA
MSMEG_3336|M.smegmatis_MC2_155 FDDLVAQLPAD-LRVVAVDLAEHG--DSRSSRTVWTMEEFARDVDAVLSA
: : * : * * * .: .
Mflv_1348|M.gilvum_PYR-GCK RGID-TAHLVGLSLGGMTAMRVAARN--PDRVNRMVLLCTAA--------
TH_3727|M.thermoresistible__bu LGLR-DAVHVGHSTGGGVVVRYLARHG-EERTAKAVLIAAVP--------
MLBr_00862|M.leprae_Br4923 LSPDRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYI
MMAR_3948|M.marinum_M LGVD-RFIFVAEGQGAVVALRTALLAP--QRVRALALIGPTA--------
MAB_0303|M.abscessus_ATCC_1997 LEVD-SFVPIAHAHGGWAALEIADRLG-AQRVPAVMILDLIM--------
MSMEG_3336|M.smegmatis_MC2_155 ESIT-RCVVVGHSLGGAVAVEVGRQLP--DVVTHVVVLDALHYLG-----
:. *. . . : . :
Mflv_1348|M.gilvum_PYR-GCK -----------------------------------ELAP----------A
TH_3727|M.thermoresistible__bu -----------------------------------PLMVRTDANPGGLPK
MLBr_00862|M.leprae_Br4923 LSGLRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRR
MMAR_3948|M.marinum_M -------------------------------------------------E
MAB_0303|M.abscessus_ATCC_1997 -------------------------------------------------T
MSMEG_3336|M.smegmatis_MC2_155 ------------------------------------------------LF
Mflv_1348|M.gilvum_PYR-GCK QAWT-------DRAAAVRAGGAESVAPAVIARWFTLS-------------
TH_3727|M.thermoresistible__bu SVFD-------DFQAQLAANRSEFYRALASGPFYGFN-------------
MLBr_00862|M.leprae_Br4923 TMVDNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNV
MMAR_3948|M.marinum_M AALD-------GENAALTAAMEIWCTQGPLPDIYRPT-------------
MAB_0303|M.abscessus_ATCC_1997 PAPR-------EFVAALHGIQDPERWKEGRDGLVQSW-------------
MSMEG_3336|M.smegmatis_MC2_155 PAMDD------SKAEAIIRPFHDDFAAAVRALVEGGS-------------
:
Mflv_1348|M.gilvum_PYR-GCK --------------HLETHPAER--AEWVDMIAAT---------------
TH_3727|M.thermoresistible__bu --------------RDGVEPSEALIANWWRQGMMG---------------
MLBr_00862|M.leprae_Br4923 PVQLIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAA
MMAR_3948|M.marinum_M --------------AAMATPTPDAADALMERWRCS---------------
MAB_0303|M.abscessus_ATCC_1997 --------------LAGTTNQAVLDHVRYDSGGHG---------------
MSMEG_3336|M.smegmatis_MC2_155 ----------------PEGFSEELIDSYFSMMVKVR--------------
Mflv_1348|M.gilvum_PYR-GCK ------------PAEGYASCCEAIA-ELDLREQLPTITAPTLAIAGADDP
TH_3727|M.thermoresistible__bu ------------DAKAHYDGIVAFS-QTDFTEDLTKITVPVLVMHGDDDQ
MLBr_00862|M.leprae_Br4923 VHDLADLVDGKQPSRALLRAQVGRS-RDTFGDTLVSVTGAGSGIGRETAL
MMAR_3948|M.marinum_M ------------AWREYRAAADALATRPTFIAELSTVACPALVVHSTQDV
MAB_0303|M.abscessus_ATCC_1997 ---------FDMWARAGRVIDEAYRTWGSPMRRMEALAEPCAIRHVFSHP
MSMEG_3336|M.smegmatis_MC2_155 -------------QPAGIRAIEGLV-RWNMEHALSETTQPITAFAVR---
. : : .
Mflv_1348|M.gilvum_PYR-GCK ATPPAKLEQIVSAIPG-ARLLVVDGAAHL---ANAEQPEQITPAIIEHLE
TH_3727|M.thermoresistible__bu VVPYANSGPLTAELLRNAKLKTYQGFPHG---MMTTHAEVVNADILEFLR
MLBr_00862|M.leprae_Br4923 ALARRGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFA
MMAR_3948|M.marinum_M FVPMFLGKEVADSLGGPTAFVPVDTGHHAITCAFHPQIGAEILAWLPSVE
MAB_0303|M.abscessus_ATCC_1997 KIG--EYDALHDDFAARHPWFSYRRLGGETHFPGIELPQQVAAEAIDLLA
MSMEG_3336|M.smegmatis_MC2_155 ---ELLSSEAVERFGDRIDFVPVELGSHHFPVESPEDTAKLLTSVVSV--
: :
Mflv_1348|M.gilvum_PYR-GCK QR------------------------------------------------
TH_3727|M.thermoresistible__bu S-------------------------------------------------
MLBr_00862|M.leprae_Br4923 EQVSAKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRA
MMAR_3948|M.marinum_M DHPE----------------------------------------------
MAB_0303|M.abscessus_ATCC_1997 GARI----------------------------------------------
MSMEG_3336|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1348|M.gilvum_PYR-GCK --------------------------------------------------
TH_3727|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 FGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAEL
MMAR_3948|M.marinum_M --------------------------------------------------
MAB_0303|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3336|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1348|M.gilvum_PYR-GCK --------------------------------------------------
TH_3727|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DAVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRLRR
MMAR_3948|M.marinum_M --------------------------------------------------
MAB_0303|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3336|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1348|M.gilvum_PYR-GCK --------------------------------------------------
TH_3727|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 YGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTARIRVI
MMAR_3948|M.marinum_M --------------------------------------------------
MAB_0303|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3336|M.smegmatis_MC2_155 --------------------------------------------------