For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDKYPVIVWGTGVVGKLVIRELLDHPVFELAAVLVHDPAKDGVDVGTLVGSHPTGLAATTDIDAALGTEG TVAYFGPTAQYALENIDNMSRALRSGHNIVSTAMTPWVYPQVCPPEMLADIQHACEAGQTSCFTTGIDPG FANDLFPMTLLGVGGRVDSVLVQELLDYQYYNGDFSTPMGLGAPMDQPAVLEIPEVLILAWGHTIPMIAD AVGVKLDKIDTVYEKWATPTEISYGSAPNTGTIPAGHCAAVRFEIRGWVGGEPKIVIEHVNRITNDTAPQ WPRAQSVDNDAYRIEIKGSPNITQETVFRDERSGDGAIGGCLATGMRAINAIPAVIDAKPGFLTPLDLPL IAGRGTIRI*
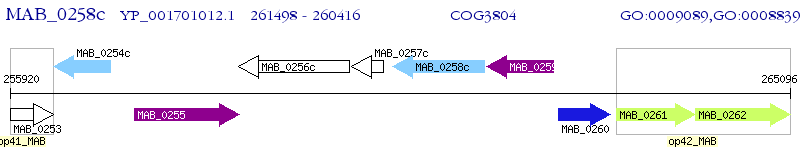
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0258c | - | - | 100% (360) | putative dihydropicolinate reductase |
| M. abscessus ATCC 19977 | MAB_4633 | - | 1e-60 | 39.11% (358) | hypothetical protein MAB_4633 |
| M. abscessus ATCC 19977 | MAB_0727 | - | 4e-30 | 29.75% (363) | putative dihydrodipicolinate reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0949c | - | 9e-65 | 40.73% (356) | hypothetical protein Mb0949c |
| M. gilvum PYR-GCK | Mflv_2763 | - | 5e-98 | 50.27% (364) | dihydrodipicolinate reductase |
| M. tuberculosis H37Rv | Rv0926c | - | 9e-65 | 40.73% (356) | hypothetical protein Rv0926c |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_3359 | - | 1e-101 | 52.65% (359) | hypothetical protein MMAR_3359 |
| M. avium 104 | MAV_2176 | dapB | 1e-103 | 53.48% (359) | dihydrodipicolinate reductase N-terminus domain-containing |
| M. smegmatis MC2 155 | MSMEG_4350 | dapB | 1e-101 | 52.49% (362) | dihydrodipicolinate reductase |
| M. thermoresistible (build 8) | TH_4297 | - | 9e-70 | 40.40% (354) | PUTATIVE putative dihydrodipicolinate reductase, N-terminus |
| M. ulcerans Agy99 | MUL_4353 | - | 1e-35 | 31.77% (362) | hypothetical protein MUL_4353 |
| M. vanbaalenii PYR-1 | Mvan_3771 | - | 1e-101 | 50.96% (365) | dihydrodipicolinate reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0949c|M.bovis_AF2122/97 ---------------------------------MAIPVVQLGTGNVGVHS
Rv0926c|M.tuberculosis_H37Rv ---------------------------------MAIPVVQLGTGNVGVHS
TH_4297|M.thermoresistible__bu -------------------------------------VILWGPGQVGVGA
Mflv_2763|M.gilvum_PYR-GCK ----MVG-------------------------SDVKRVVIWGTGFVGRMV
Mvan_3771|M.vanbaalenii_PYR-1 ----MACGAASEM-------------------TDVKRVVIWGTGFVGRMV
MMAR_3359|M.marinum_M ----MSQ-------------------------SGGKRVVVWGTGFVGRMV
MAV_2176|M.avium_104 MAARMSE-------------------------HGSKRVVVWGTGFVGKMV
MSMEG_4350|M.smegmatis_MC2_155 ----MTGAAPVLGGTLHVPNPIPGVTGITYRGRVKKRVVVWGTGFVGSMV
MAB_0258c|M.abscessus_ATCC_199 -------------------------------MSDKYPVIVWGTGVVGKLV
MUL_4353|M.ulcerans_Agy99 -----------------------------MSGPAKLRVIQWATGGVGKAA
*: ..* **
Mb0949c|M.bovis_AF2122/97 LRALIADPEFELTGVWVSSDAKAGKDAAELAGLADSTGVRASTDLNAVLA
Rv0926c|M.tuberculosis_H37Rv LRALIADPEFELTGVWVSSDAKAGKDAAELAGLADSTGVRASTDLNAVLA
TH_4297|M.thermoresistible__bu LRAVIAHPGLELVGLVVHSPAKHGKDAGALCGMP-ETGILATNDIDAALA
Mflv_2763|M.gilvum_PYR-GCK IAEVVGHPRFELVGVGVSNPDKVGRDVGDICGLGRDLGLTATDDVDALID
Mvan_3771|M.vanbaalenii_PYR-1 IPEVVRHPQFELVGVGVSNPDKAGRDVGEICGMGRDIGLIATDDVDALID
MMAR_3359|M.marinum_M IPEIVKHPLFELVGVGVSNPDKVGRDVGEICGLSEPVGISATDDVDALIA
MAV_2176|M.avium_104 IAEIVKHPLFELVGVGVSNPAKVGRDVADICGLPEPTGVIATDDVDALIA
MSMEG_4350|M.smegmatis_MC2_155 IAEIVGHPLFELVGVGVSNPDKVGRDVGEICGLGAPVGVTATDDVDALIA
MAB_0258c|M.abscessus_ATCC_199 IRELLDHPVFELAAVLVHDPAKDGVDVGTLVGS-HPTGLAATTDIDAALG
MUL_4353|M.ulcerans_Agy99 IECILNHPQLELAGCWVHSAAKHGLDVGTIIGAA-RFGVTATNDVDELLA
: :: .* :**.. * . * * *.. : * *: *: *:: :
Mb0949c|M.bovis_AF2122/97 TGPRCAVYNAMADNRLPEALEDYRRILAAGINIVGSGPVFLQYPW-QVIP
Rv0926c|M.tuberculosis_H37Rv TGPRCAVYNAMADNRLPEALEDYRRILAAGINIVGSGPVFLQYPW-QVIP
TH_4297|M.thermoresistible__bu VDADVVAYFASGDYRYREAAEDIARCLRAGKNVVTTSLVPLCYP--PAAD
Mflv_2763|M.gilvum_PYR-GCK LKPDALVHYGPTAARADANIELITRFLRAGIDVCSTAMTPWIWPTMHLNP
Mvan_3771|M.vanbaalenii_PYR-1 LKPDALVHYGPTAAHAEANIELITRFLRAGIDVCSTAMTPWIWPTMHLNP
MMAR_3359|M.marinum_M LRPDALVHYGPTAMHAQDNISLITRFLRAGIDVCSTAMTPWIWPTMHLNP
MAV_2176|M.avium_104 LKPDALVHYGPTAMHAKENIALITRFLRAGIDVCSTAMTPWVWPTMHLNP
MSMEG_4350|M.smegmatis_MC2_155 LKPDALVHYGPTAAQADANIALITRFLRAGIDVCSTAMTPWVWPKMHLNP
MAB_0258c|M.abscessus_ATCC_199 TEG-TVAYFGPTAQYALENIDNMSRALRSGHNIVSTAMTPWVYP--QVCP
MUL_4353|M.ulcerans_Agy99 LDADCVMYSPQVAD-----EQDVKAILRSGKNVFTP--VGWVYP---DLS
: * :* :: . . :*
Mb0949c|M.bovis_AF2122/97 DEIIKPLQDAARAGNSSLYVNGIDPGFANDLLPMALAGTCESIEQIRCME
Rv0926c|M.tuberculosis_H37Rv DEIIKPLQDAARAGNSSLYVNGIDPGFANDLLPMALAGTCESIEQIRCME
TH_4297|M.thermoresistible__bu AETVDLIEAACAEGRTTFFNSGVDPGWANDLMALTMTGFSSRVDTITMLE
Mflv_2763|M.gilvum_PYR-GCK PNWIEPVTVACELGESSCFTTGIDPGFANDLFPMTLMGLCSEVRTVRASE
Mvan_3771|M.vanbaalenii_PYR-1 PNWIEPITVACELGESSCFTTGIDPGFANDLFPMTLMGLCSEVRRVRASE
MMAR_3359|M.marinum_M PQWIQPITEACTLGESSCFTTGIDPGFANDLFPMTLMGLCSEVQRVRASE
MAV_2176|M.avium_104 PNWIAPITEACELGESSCFTTGIDPGFANDLFPMTLMGLCSEVRRVRASE
MSMEG_4350|M.smegmatis_MC2_155 PNWIEPITEACEVGQSSCFTTGIDPGFANDLFPMTLMGLCSEVRRVRASE
MAB_0258c|M.abscessus_ATCC_199 PEMLADIQHACEAGQTSCFTTGIDPGFANDLFPMTLLGVGGRVDSVLVQE
MUL_4353|M.ulcerans_Agy99 KPRVKAVADAAVAGGVTLHGSGIHPGGITERFPLMVSSLSSAITHVRAEE
: : *. * : . .*:.** .: :.: : . : : *
Mb0949c|M.bovis_AF2122/97 IVDYATYDSAVVMFDVMGFGKPMDQIPMLLQPGVLSLAWGSVVRQLAAGL
Rv0926c|M.tuberculosis_H37Rv IVDYATYDSAVVMFDVMGFGKPMDQIPMLLQPGVLSLAWGSVVRQLAAGL
TH_4297|M.thermoresistible__bu ILDYAPINQPEIMFDFMGFGHPPEHPAPLFDPERLAALWAPTVHLVADGV
Mflv_2763|M.gilvum_PYR-GCK LLDYTNYEG--DYDREMGIGKPPEFTPMLENPDILVFAWGATVPMIAHAA
Mvan_3771|M.vanbaalenii_PYR-1 LLDYTNYEG--DYDREMGIGRPPEYRPMLENPDILVFAWGATVPMIAHAA
MMAR_3359|M.marinum_M LLDYTNYEG--DYEKEMGIGREPEFSPMLENRDVLIFAWGATVPMIAHAA
MAV_2176|M.avium_104 LLDYTNYEG--DYEVEMGIGREPEYSPMLENRDVLIFAWGATVPMIAHAA
MSMEG_4350|M.smegmatis_MC2_155 LLDYTNYTG--DYEFEMGIGKPPEHRPLLESPDILIFAWGATVPMIAHAA
MAB_0258c|M.abscessus_ATCC_199 LLDYQYYNG--DFSTPMGLGAPMDQPAVLEIPEVLILAWGHTIPMIADAV
MUL_4353|M.ulcerans_Agy99 FSDIRTYNAPDVVRDIMGFGVPHAQAMQGPMAALLESGFKMSVRMIANHM
: * **:* * : : :*
Mb0949c|M.bovis_AF2122/97 GISLDGVEEMYVREPAPEAFNI----ASGHIPKGSAAALRFEVLGLVDGV
Rv0926c|M.tuberculosis_H37Rv GISLDGVEEMYVREPAPEAFNI----ASGHIPKGSAAALRFEVLGLVDGV
TH_4297|M.thermoresistible__bu GLPLDRVETTIDKWLATERYRV----ASGWIEPGTMGGMRFKLAGIVDGE
Mflv_2763|M.gilvum_PYR-GCK GIMLDEITTTWDKWVTPDERRT----AKGVIAPGNVAAVRFTINGVYRGE
Mvan_3771|M.vanbaalenii_PYR-1 GIMLDEITTTWDKWVTPTERKT----AKGVIAPGHVAAVRFTVNGVYRGE
MMAR_3359|M.marinum_M GIMLDEITTTWEKWVTPTERKS----AKGLIKAGHVAAVRFTINGVYQGE
MAV_2176|M.avium_104 GIMLDEITTTWDKWVTPTERTT----AKGVIKPGHVAAVRFTINGVYRGE
MSMEG_4350|M.smegmatis_MC2_155 GVELDEITTTWDKWVTPDDRKT----AKGVIGAGNVAAVRFTINGVYRGE
MAB_0258c|M.abscessus_ATCC_199 GVKLDKIDTVYEKWATPTEISYGSAPNTGTIPAGHCAAVRFEIRGWVGGE
MUL_4353|M.ulcerans_Agy99 GFHIDPMIRATHEIAVATSDIAS---DVLPIAADTVAGRKFRWQTLADGK
*. :* : . .. * . .. :* *
Mb0949c|M.bovis_AF2122/97 PAVVLEHVTRLR-ADLCPEWPQPAQ-PGGSYRIEISGEPCYAMDI-CLSS
Rv0926c|M.tuberculosis_H37Rv PAVVLEHVTRLR-ADLCPEWPQPAQ-PGGSYRIEISGEPCYAMDI-CLSS
TH_4297|M.thermoresistible__bu PRVVLEHITRMG-EHSAPDWPRHPS-PHGGYRVVVDGLPTYTVDI-EMHG
Mflv_2763|M.gilvum_PYR-GCK TRIQLEHVNRIG-HDAAPDWPSGT--ESDVYRVDIEGTPSIFQET-AFRF
Mvan_3771|M.vanbaalenii_PYR-1 TRIQLEHVNRIG-SDAAPDWPSGN--ENDVYRVDIEGTPSIFQET-AFRF
MMAR_3359|M.marinum_M TRIQLEHVNRIG-HDAAPDWPSGH--DDDVYRVDIEGTPSIFQET-AFRF
MAV_2176|M.avium_104 TRIQLEHVNRIG-LDAAPDWPSGH--DNDVYRVDIEGTPSIFQET-AFRF
MSMEG_4350|M.smegmatis_MC2_155 TRIQLEHVNRIG-ADAAPDWPTGN--DNDVYRVDIEGTPSISQET-AFRF
MAB_0258c|M.abscessus_ATCC_199 PKIVIEHVNRIT-NDTAPQWPRAQSVDNDAYRIEIKGSPNITQET-VFR-
MUL_4353|M.ulcerans_Agy99 PVITAAVNWLMGQADMHPDWSFGER--GERFEIEITGNPSVSLVFKGLQP
. : : . *:*. :.: : * * :
Mb0949c|M.bovis_AF2122/97 RHGDHNHAGLV---ATAMRIVNAIPAVVAAEPGIRTTLDLPLITG---EG
Rv0926c|M.tuberculosis_H37Rv RHGDHNHAGLV---ATAMRIVNAIPAVVAAEPGIRTTLDLPLITG---EG
TH_4297|M.thermoresistible__bu R-GNNMRGLTY---ATVMRELNAIPAVVAAPPGLLSTLDLPLVTGPVRGG
Mflv_2763|M.gilvum_PYR-GCK TDGSGRDAAAAGCLATGLRALNAVPAVNDLSPGWVTALDLPLIPG---VG
Mvan_3771|M.vanbaalenii_PYR-1 TDGSGRDAAAAGCLATGLRALNAVPAVNDLSPGWVTALDLPLIPG---LG
MMAR_3359|M.marinum_M TDGSGRDPATAGCLATGMRALNAVPAVNDLPPGWVTALDLPLIPG---AG
MAV_2176|M.avium_104 TDGSGRDAAAAGCLATGLRALNAVPAVNELRPGWVTPLDLPLIPG---TG
MSMEG_4350|M.smegmatis_MC2_155 TDGSGRDAAAAGCLATGLRALNAVPAVNQHLPGWVTPLDLPLIPG---LG
MAB_0258c|M.abscessus_ATCC_199 DERSG-DGAIGGCLATGMRAINAIPAVIDAKPGFLTPLDLPLIAG---RG
MUL_4353|M.ulcerans_Agy99 TSITEGLQRNPGVVATANHCANAIPVVCAADPGIATYLDLPLFAGRPARA
** : **:*.* ** : *****..* .
Mb0949c|M.bovis_AF2122/97 RYAAA-------
Rv0926c|M.tuberculosis_H37Rv RYAAA-------
TH_4297|M.thermoresistible__bu RWTGTLPPTTRR
Mflv_2763|M.gilvum_PYR-GCK TIR---------
Mvan_3771|M.vanbaalenii_PYR-1 TIR---------
MMAR_3359|M.marinum_M TIR---------
MAV_2176|M.avium_104 TIR---------
MSMEG_4350|M.smegmatis_MC2_155 TIR---------
MAB_0258c|M.abscessus_ATCC_199 TIRI--------
MUL_4353|M.ulcerans_Agy99 SRGLER------